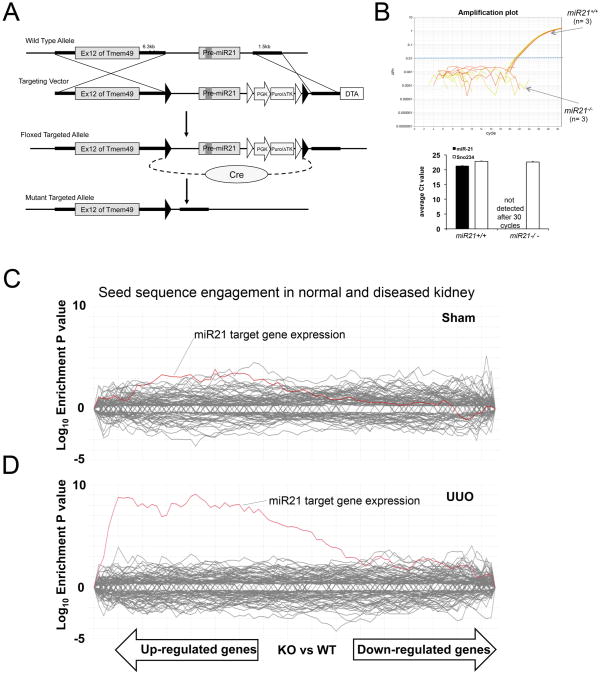
Fig. 2.
Development and characterization of miR-21 targeted mutation in mice. (A) Schema showing targeting strategy for miR-21 in intron 12 of the Tmem49 gene. LoxP sites [black triangles] were inserted around Pre-miR-21 and the inserted puromycin resistance gene. Cre mediated excision was performed in ES cells prior to generation of mutant mice. (B) Taqman Amplification plot (upper) showing amplification of miR-21 in RNA extracted from miR21+/+ kidney but not miR21-/- kidney, and graph (lower) showing expression level of mature miR-21 from miR-21+/+ and miR-21-/- kidneys. The small nucleolar RNA Sno234 was used as a control. (C-D) SylArray analyses (16) showing whole kidney microarray data [30,000 genes] from miR-21-/- compared with miR-21+/+ kidneys. Each line shows the relative expression intensity [miR-21-/- relative to miR-21+/+] for all mRNA transcripts that contain a target (seed) sequence for a particular microRNA in their 3′ untranslated region. The red line identifies all mRNA transcripts with seed sequence for miR-21. In normal miR-21-/- kidney (C), there is no significant enrichment of miR-21 target genes, but in miR-21-/- kidney d7 after UUO (D) there is marked and highly significant enrichment of expression of miR-21 target gene transcripts in the microarray.