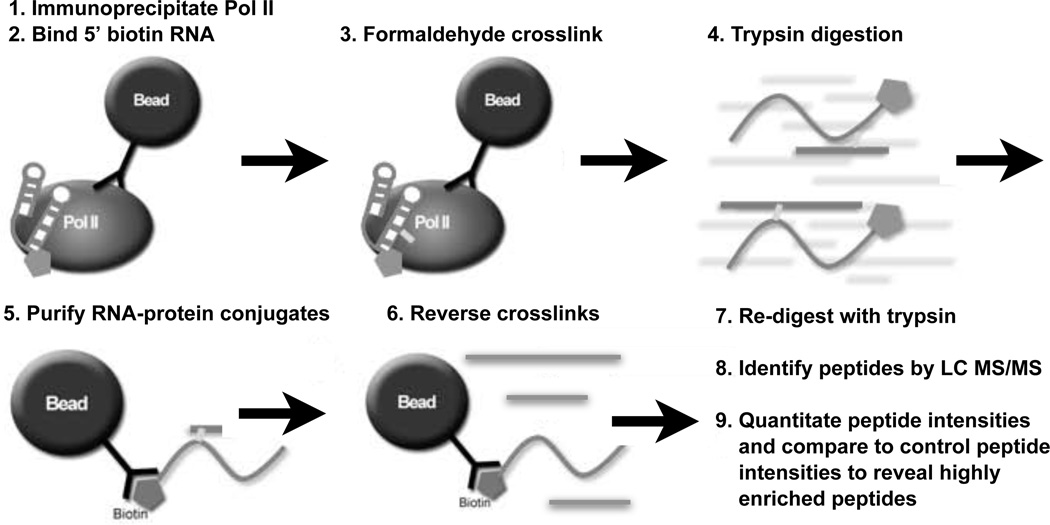
Figure 3.
Schematic overview of the ncRNA-Pol II formaldehyde crosslinking protocol. (1) Pol II was immunoprecipitated from nuclear extracts using bead-immobilized antibody. (2) 5'-biotinylated RNA was bound to Pol II, followed by washes with a nonspecific competitor DNA and higher salt to reduce nonspecific binding. (3) Complexes were treated with formaldehyde to covalently crosslink ncRNA to Pol II. (4) The entire solution containing the RNA and bead-bound Pol II was digested with trypsin. (5) After centrifugation, the supernatant was mixed with streptavidin beads to purify the ncRNA and enrich for RNA-crosslinked peptides. (6) After centrifugation the supernatant was heated to reverse formaldehyde crosslinks. (7) Peptides were then re-digested with trypsin. (8) Recovered peptides were subjected to LC MS/MS sequencing and identified using Mascot. (9) The intensities of peptides recovered from the crosslinked samples were compared to those from the control samples as described in the text.