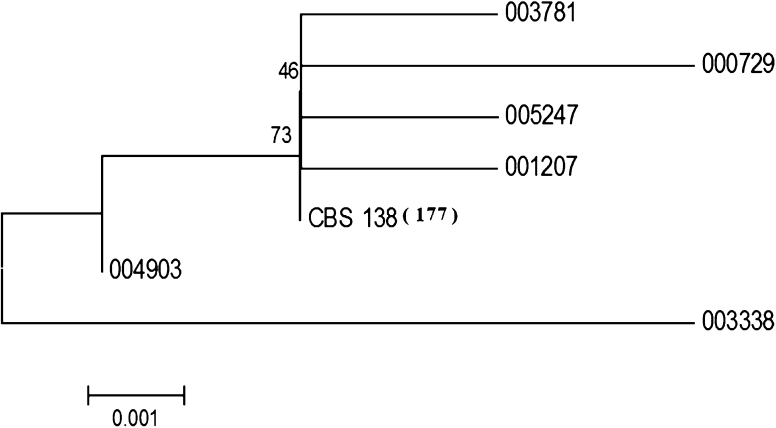
Fig. 1.
Phylogenetic relationships among pathogenic C. glabrata strains, based on seven different haplotypes, as deduced by Neighbor-joining method. The analysis is based on the D1/D2 domain of the 26 rDNA encoding locus. The numbers correspond to the museum numbers of the initial collection and can be found in Supplementary materials Table S1. Among the analyzed sequences (for accession numbers see Table S1), which had the 489 positions, 177 were identical with the C. glabrata type strain CBS 138. The strains belonging to the same haplotype are described in Supplementary materials Table 1. The bootstrap values are shown on some branches and the tree was not rooted. The scale bar in Neigbour-joining analysis corresponds to 0.001substitution per nucleotide site