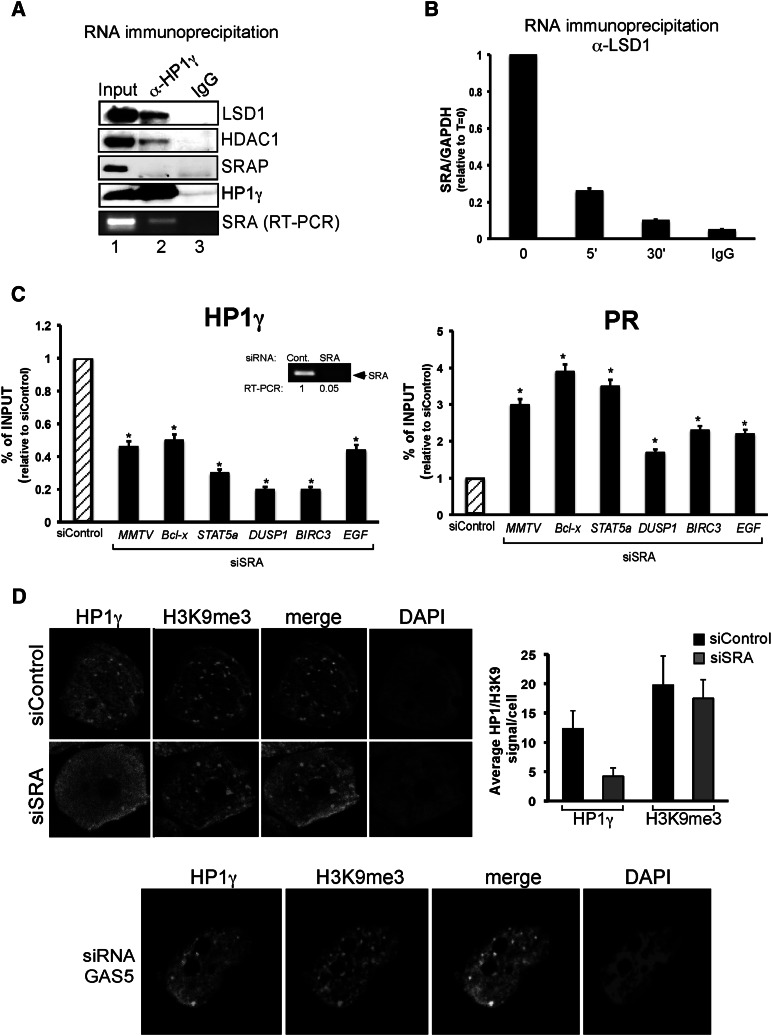
Figure 5.
The SRA RNA is involved in repressive complex anchoring to chromatin. (A) T47D-MTVL cells were lysed and immunoprecipitated with either α-HP1γ-specific antibody or mouse IgG and separated into two fractions for RNA and Western blot analysis. (Bottom panel) Either the immunoprecipitates were analyzed by immunoblotting with LSD1-, HDAC1-, SRAP-, and HP1γ-specific antibodies or total RNA was extracted and subjected to RT–PCR with the primers specific to SRA. (B) Cells were treated with hormone for the indicated times and immunoprecipitated with either α-LSD1-specific antibody or rabbit IgG, and total RNA was extracted and subjected to RT–PCR with the primers specific to SRA and GAPDH. The values represent the mean and standard deviation from three experiments performed in duplicate. (C) Cells were transfected with either control or SRA-specific siRNAs and subjected to ChIP assays using HP1γ and PR antibodies. DNA was analyzed for the presence of MMTV nucleosome B, BCL-X, DUSP1, STAT5A, BIRC3, and EGF PRbs. The values represent the mean and standard deviation from three experiments performed in duplicate. (*) P-value < 0.05. The degree of depletion of SRA RNA is depicted. (D) Cells were transfected with either control, SRA-specific (top panels) or GAS5-specific (bottom panel) siRNAs; fixed; and immunostained with HP1γ and H3K9me3-specific antibodies or DAPI. (Right panel) T47D-MTVL cells were transfected with the indicated siRNA, and the number of HP1 or H3K9me3 dots per cell was counted using the ImageJ software for three different images per condition.