Figure 5.
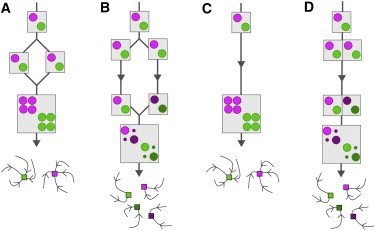
Evolutionary scenarios for the simultaneous duplication and divergence of multiple genes. We start with a cell that contains two distinct weakly interacting sets of coats and SNAREs (represented by pictorial matrices, as in Fig. 4), and therefore have two distinct organelles (represented as schematic compartment trajectories in compositional space). (A and B) Hybridization scenarios. (A) If two identical daughter cells of a single mother fuse, the new protein copies will be identical to the old copies, so the interaction matrix will have two blocks. The result is a two-organelle steady state. (B) If the daughter cells are allowed to diverge over evolutionary timescales, protein complexes within a single lineage will coevolve, but will tend to lose interactions with protein complexes in the other lineage. If these diverged cells eventually fuse, their interactions will have the correct diagonal form. The result is a four-organelle steady state. (C and D) Neolocalization scenarios. (C) We start with a whole-genome duplication in which a cell fails to segregate a newly replicated genome. As in Fig. 5A, the new protein copies are identical to the old copies and the interaction matrix has two blocks. The new copies are constrained from diverging, because they are still confronted with their old interaction partners. The result is a two-organelle steady state. (D) Now we imagine that some of the new protein copies are retargeted to a different compartment of the cell and therefore never see their old partners. If these neolocalized proteins somehow contribute to fitness, they will maintain interactions among themselves, but cross-interactions with partners in other compartments will tend to be lost. If the four protein sets are eventually retargeted to the same compartment, their combined interaction matrix will have the correct diagonal form. The result is a four-organelle steady state.
