Abstract
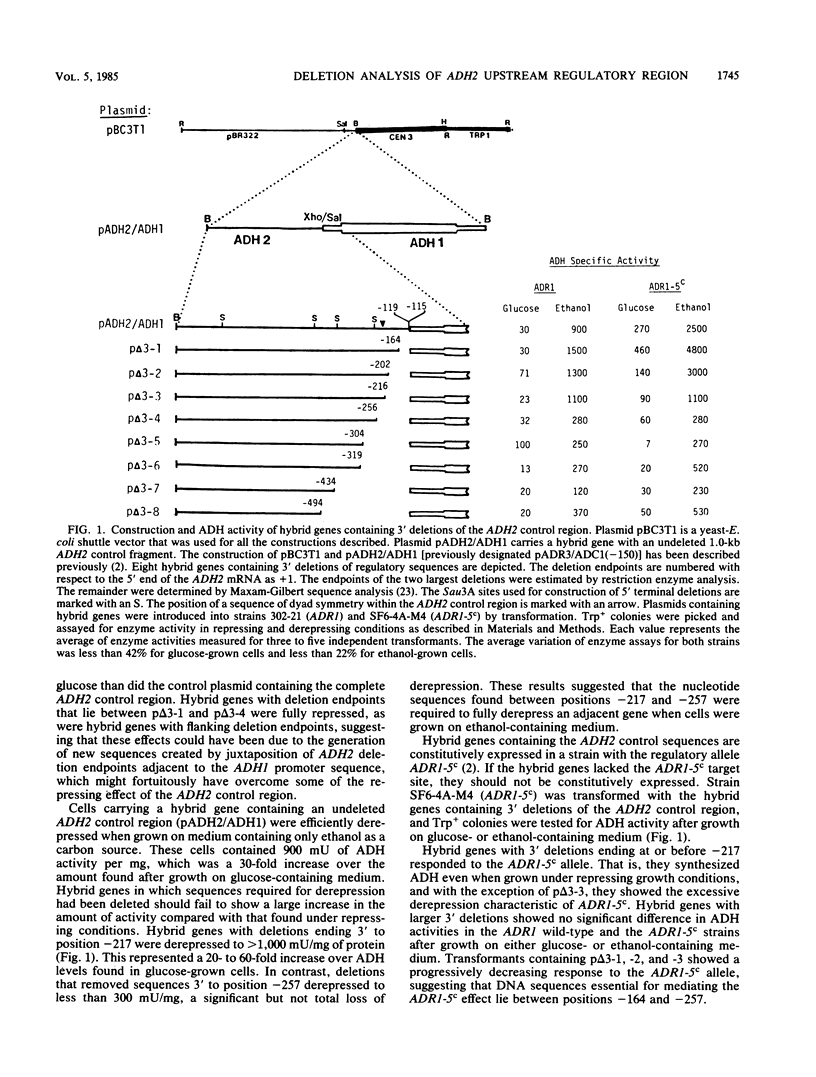
Deletion analysis was used to identify sequences upstream of the ADH2 gene of Saccharomyces cerevisiae that are required for its regulation. 5' and 3' internal deletions of the ADH2 control region were created in vitro, and the fragments were ligated adjacent to the ADH1 promoter and structural gene. Hybrid genes with 3' deletions extending from -119 to -216 (the start site of ADH2 transcription is designated +1) were fully repressed and derepressed to high levels. Hybrid genes with 3' deletions extending from -119 to -257 were repressed but failed to significantly derepress. Hybrid genes lacking the -216 to -257 region also failed to respond to ADR1-5c, a mutant allele of the unlinked regulatory gene ADR1, which confers constitutive expression on ADH2. This implies that the region between these deletion endpoints, which includes a 22-base-pair sequence of dyad symmetry, is required for efficient derepression of an adjacent promoter. Internal deletions extending in the 3' direction from position -1141 confirmed these results. Deletion mutants lacking the region -1141 to -259 were normally regulated, whereas deletions extending from -1141 to -115 were not derepressible. These results support the hypotheses that the ADH2 promoter may normally be in an inactive conformation in the yeast chromosome and that derepression of ADH2 requires positive activation mediated through an upstream activation sequence located between 216 and 257 base pairs 5' to the start site of ADH2 transcription. No evidence for a DNA sequence mediating repression was obtained.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Beier D. R., Young E. T. Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature. 1982 Dec 23;300(5894):724–728. doi: 10.1038/300724a0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chan V. L., Smith M. In vitro generation of specific deletions in DNA cloned in M13 vectors using synthetic oligodeoxyribonucleotides: mutants in the 5'-flanking region of the yeast alcohol dehydrogenase II gene. Nucleic Acids Res. 1984 Mar 12;12(5):2407–2419. doi: 10.1093/nar/12.5.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Nov;176(3):427–431. doi: 10.1007/BF00333107. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Denis C. L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984 Dec;108(4):833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Young E. T. Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Mar;3(3):360–370. doi: 10.1128/mcb.3.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Levine H. L., Goeddel D. V., Ammerer G., Hall B. D. Expression of a human gene for interferon in yeast. Nature. 1981 Oct 29;293(5835):717–722. doi: 10.1038/293717a0. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Rowe L. B. Molecular expression and regulation of the galactose pathway genes in Saccharomyces cerevisiae. Distinct messenger RNAs specified by the Gali and Gal7 genes in the Gal7-Gal10-Gal1 cluster. J Biol Chem. 1978 Oct 25;253(20):7566–7569. [PubMed] [Google Scholar]
- Lutstorf U., Megnet R. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. I. Physiological control of ADH-2 and properties of ADH-2 and ADH-4. Arch Biochem Biophys. 1968 Sep 10;126(3):933–944. doi: 10.1016/0003-9861(68)90487-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T., Oshima Y. Cyclic AMP may not be involved in catabolite repression in Saccharomyces cerevisiae: evidence from mutants unable to synthesize it. J Bacteriol. 1983 Nov;156(2):898–900. doi: 10.1128/jb.156.2.898-900.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. C., Spence A., Smith M. The distal transcription signals of the herpesvirus tk gene share a common hexanucleotide control sequence. Cell. 1984 May;37(1):253–262. doi: 10.1016/0092-8674(84)90321-0. [DOI] [PubMed] [Google Scholar]
- Perlman P. S., Mahler H. R. Derepression of mitochondria and their enzymes in yeast: regulatory aspects. Arch Biochem Biophys. 1974 May;162(1):248–271. doi: 10.1016/0003-9861(74)90125-8. [DOI] [PubMed] [Google Scholar]
- Polakis E. S., Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965 Oct;97(1):284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Position effects in Saccharomyces cerevisiae. J Mol Biol. 1981 Nov 5;152(3):569–575. doi: 10.1016/0022-2836(81)90269-2. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Bennetzen J., Young E. T., Nasmyth K., Hall B. D. Isolation of the structural gene for alcohol dehydrogenase by genetic complementation in yeast. Nature. 1980 Jan 10;283(5743):214–216. doi: 10.1038/283214a0. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Montgomery D. L., Nichols D. L., Hall B. D. Transcriptional regulation of the yeast cytochrome c gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3627–3631. doi: 10.1073/pnas.76.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]


