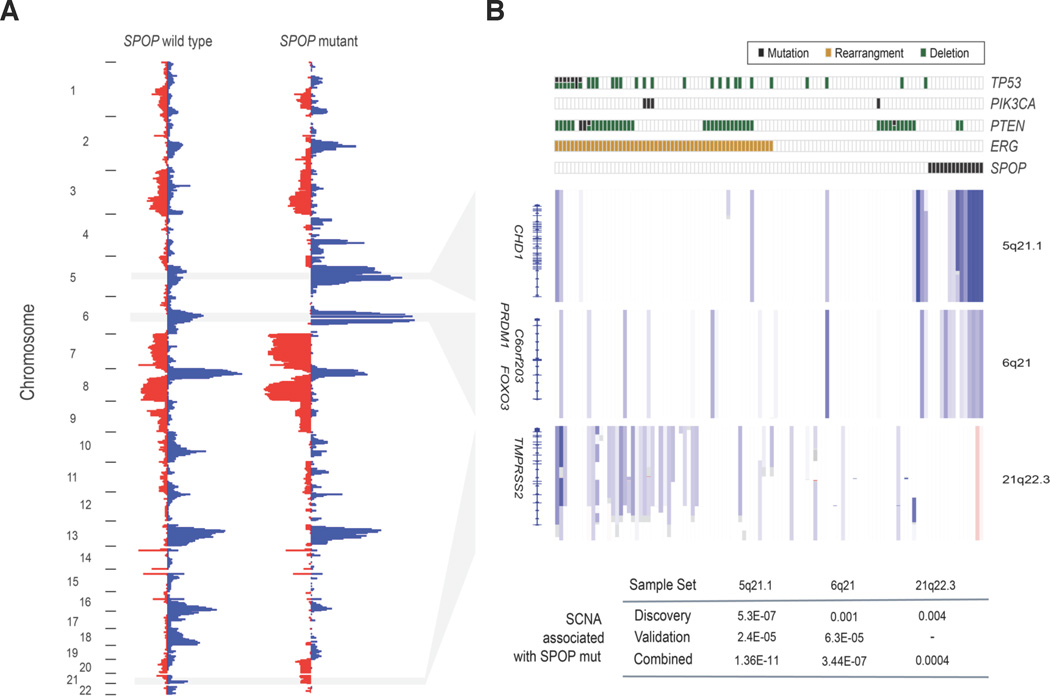
Fig 4. SPOP mutation defines a distinct genetic subclass of prostate cancer.
(Left) Frequency of genomic copy number alterations in SPOP-mutant and SPOP-wildtype tumors. Length of bars reflects the frequency of copy number loss (blue) or gain (red). (Right) Heatmap showing selected recurrent somatic copy number aberrations (SCNA). Each row represents a single prostate cancer sample. Samples are annotated for mutations in SPOP, PTEN, PIK3CA, and TP53, deletions of PTEN, and ERG rearrangements. Deletions positively correlated (5q21, 6q21) or inversely correlated (21q22.3) with SPOP mutation are shown. P-values of peak association with SPOP mutation in both discovery and validation cohorts are displayed at bottom (Fisher’s exact test). Regions are not to scale; full coordinates available in Supplementary Table 8.