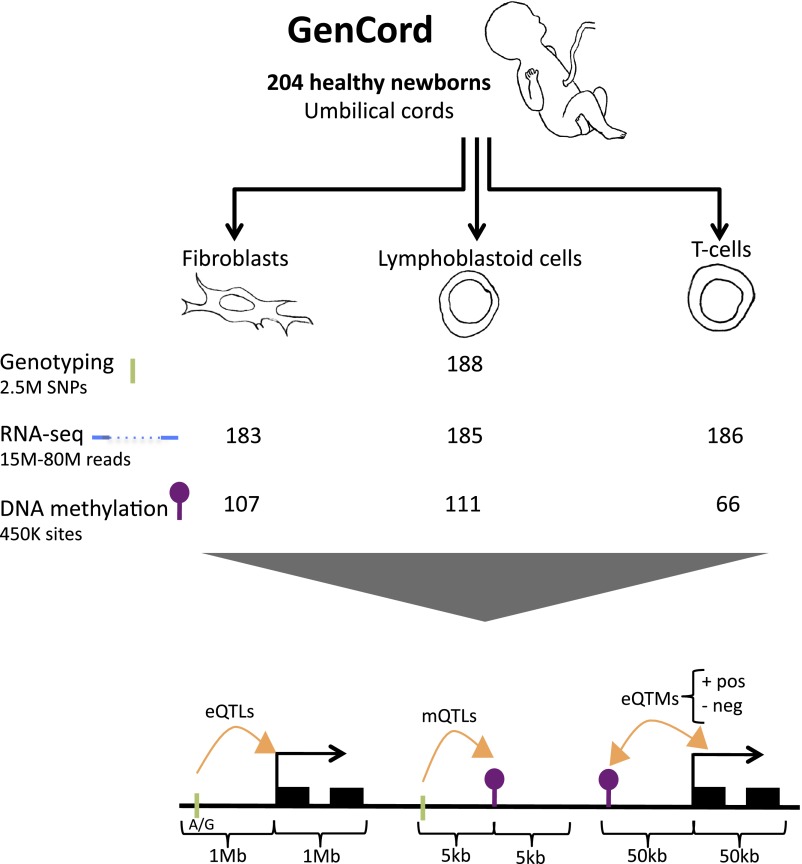
Figure 1. GenCord project scheme.
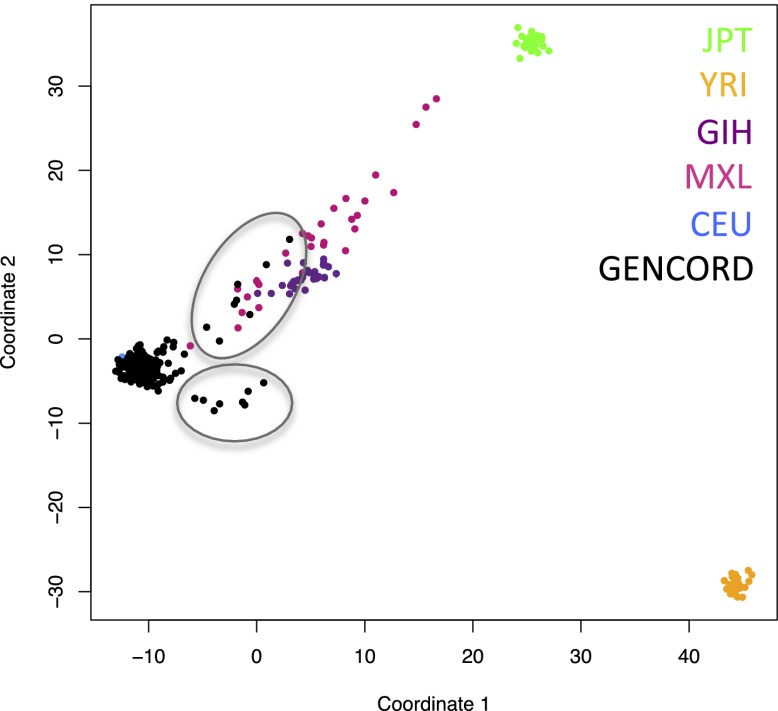
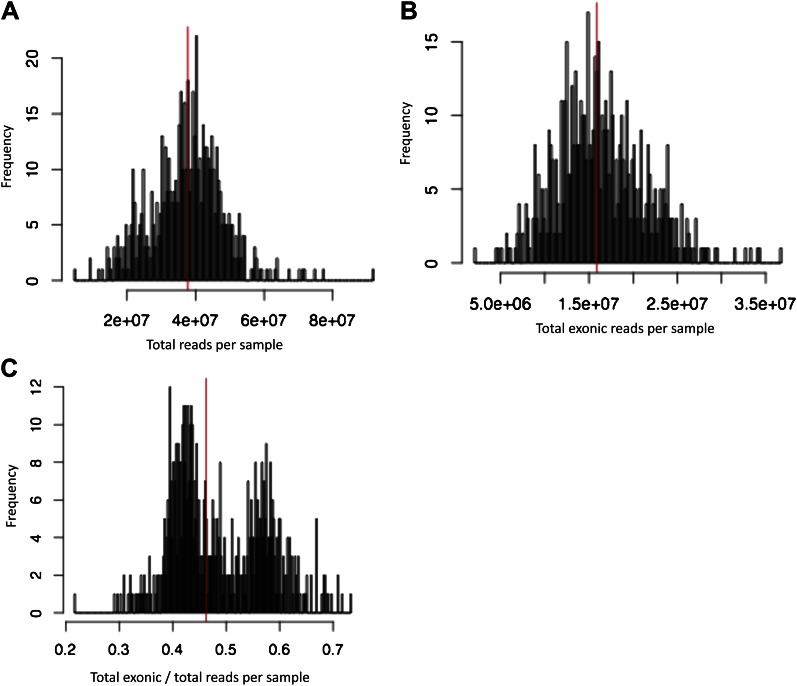
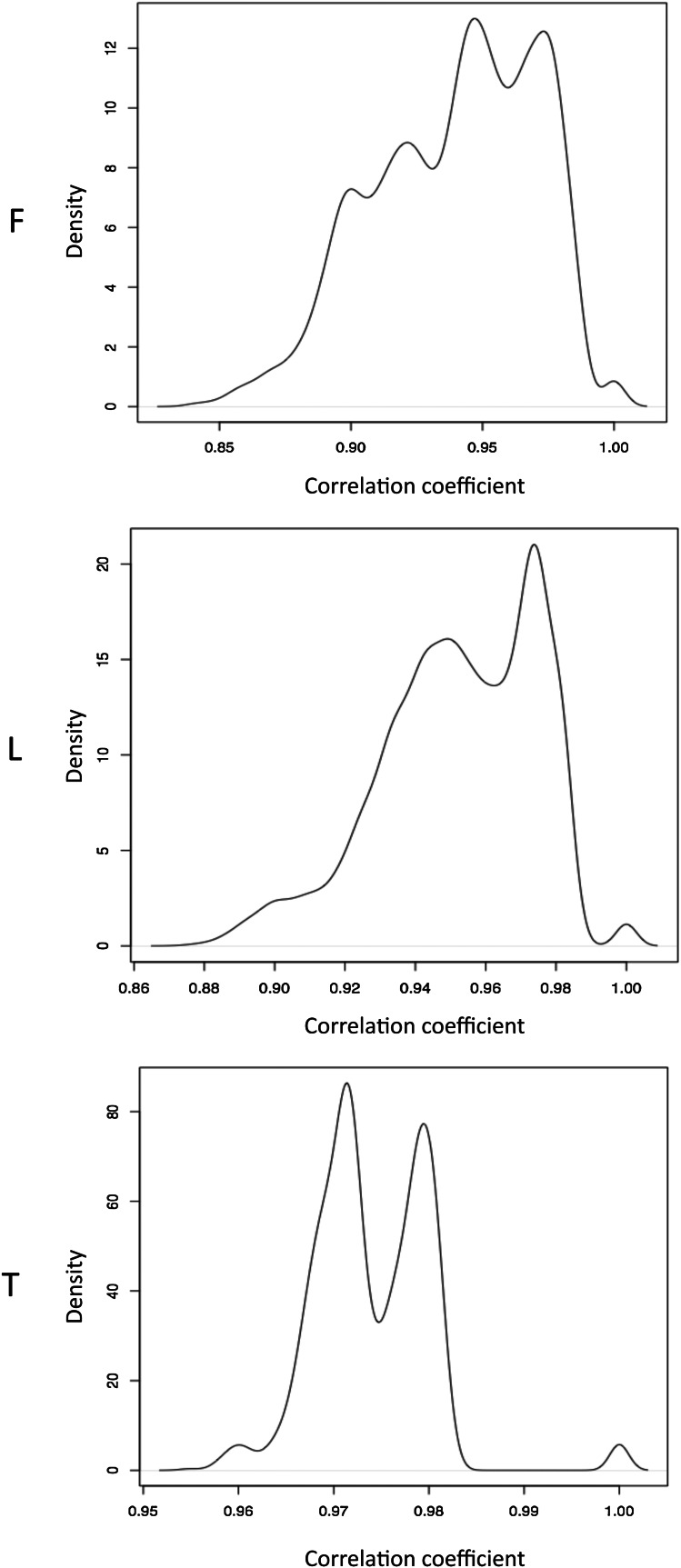
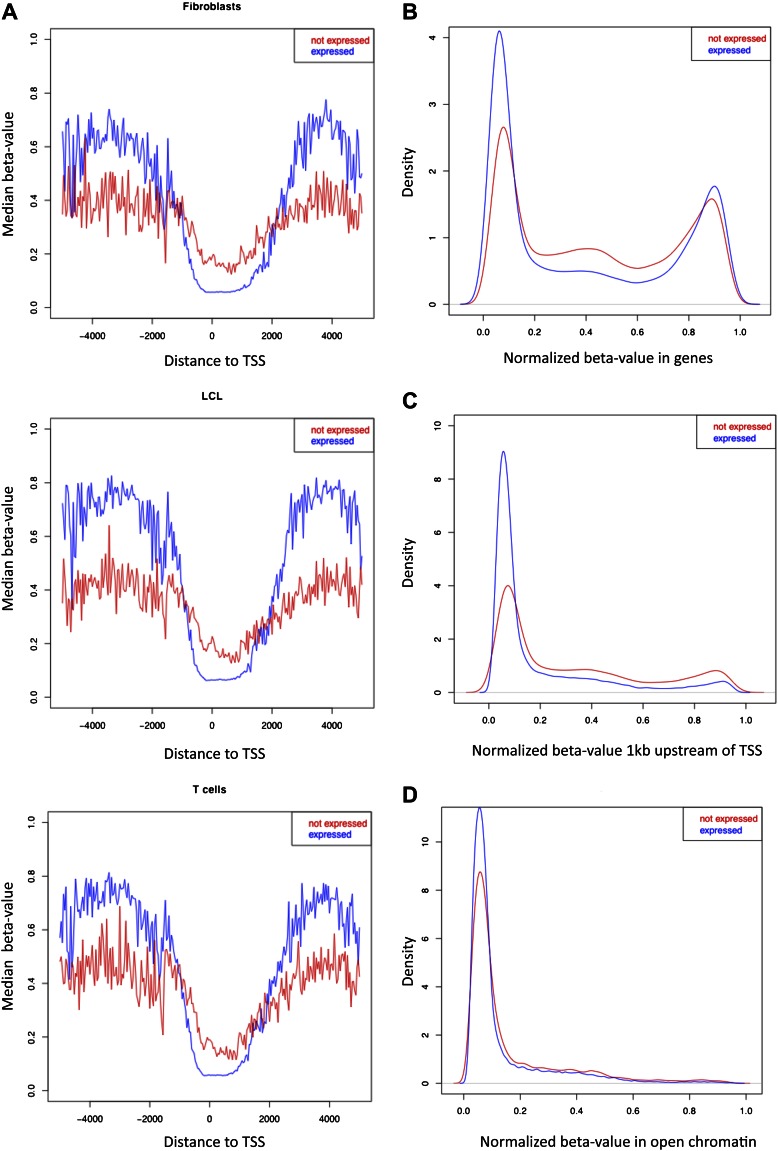
We collected umbilical cord and cord blood samples from 204 newborn babies, from which we derived three cell-types: fibroblasts, lymphoblastoid cells and T-cells. Genotyping, RNA-sequencing and DNA methylation levels were assayed. The number of samples without genetic and technical outliers is indicated for each assay and each cell-type. We then correlated and utilized different properties of all datasets in order to assess: expression Quantitative Trait Loci (eQTLs), methylation QTLs (mQTLs), positive (pos) and negative (neg) expression Quantitative Trait Methylation (eQTMs). Green ticks represent Single Nucleotide Polymorphisms (SNPs), purple lollipops represent methylation sites, black boxes represent exons and orange arrows depict associations between two data-types. Shown are the maximum distances between each pair of variables tested. See Figure 1—figure supplement 1–7 for data processing and quality checks.