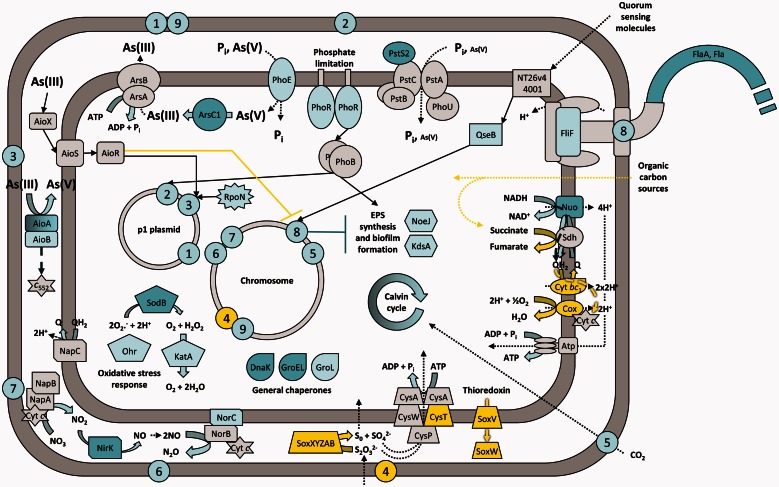
Fig. 7.—
Conceptual representation of the Rhizobium sp. NT-26 response to arsenite exposure. This representation takes into account our genomic, transcriptomic, proteomic, and physiological results as well as data from the literature. Numbers 1 to 9 represent biological functions and the approximate genomic location of the gene clusters encoding their corresponding proteins. 1 and 9: arsenate reduction; 2: phosphate and arsenate transport; 3 and 4: arsenite and sulfur oxidation, respectively; 5: carbon fixation; 6 and 7: nitrate and nitrite reduction, respectively; 8: motility. Block, dashed, dotted, and standard arrows symbolize chemical reactions, electron flow, transport/utilization of molecules and signaling/regulatory pathways, respectively. When highlighted in dark blue, light blue, yellow or gray, elements have been identified as being induced by proteomic, induced by transcriptomic, repressed by transcriptomic, and present in the genome, respectively. For clarity reasons, proteins for which the exact function is still unknown but that are related to the different processes are not shown, the protein complexes of the respiratory chain, that is the NADH dehydrogenase, the fumarate reductase, the cytochrome bc1 and the cytochrome c oxidase are designated by Nuo, Sdh, Cyt bc1, and Cox, respectively, plasmid p2 is not shown and only one flagella is represented. Finally, Cyt c and c552 are for cytochrome c and cytochrome c552, respectively, and NT26v4_4001 is a homolog of qseC.