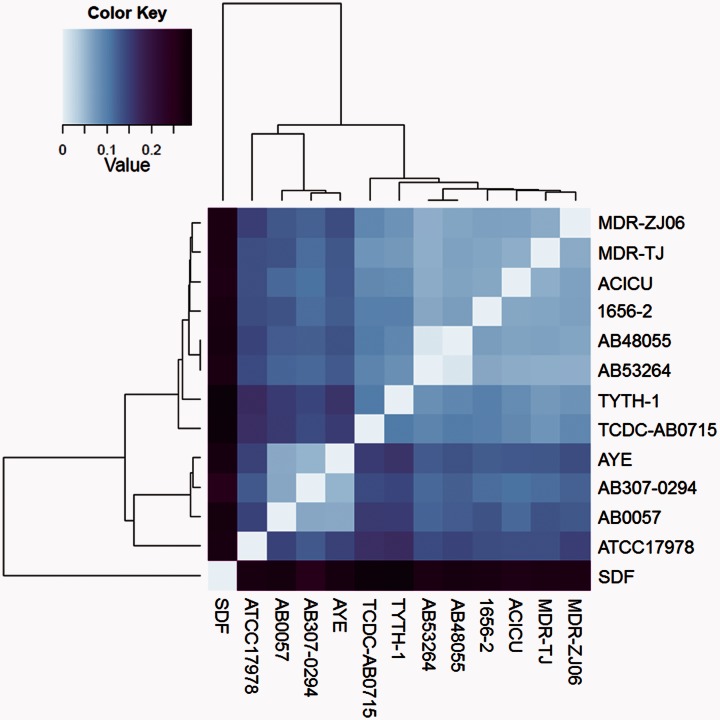
Fig. 1.—
Heat map based on a pair-wise distance matrix of whole-genome alignment as computed by Progressive Mauve. Pair-wise genome alignments were performed using the genomes of Acinetobacter baumannii 53264, A. baumannii 48055, and 11 A. baumannii clones whose complete sequences were available in the KEGG database. This heat map was created using the R statistical program (http://www.r-project.org/) with heatmap clustering methods. Dendrograms across the top and left of the diagram indicate the relatedness of the genomes based on genome conservation, while strain names are listed to the right of the heatmap. Distance values range from 0.0 to 0.3 and correspond to a gradient of color steps ranging from light blue (lowest distance value) to dark purple (highest distance value).