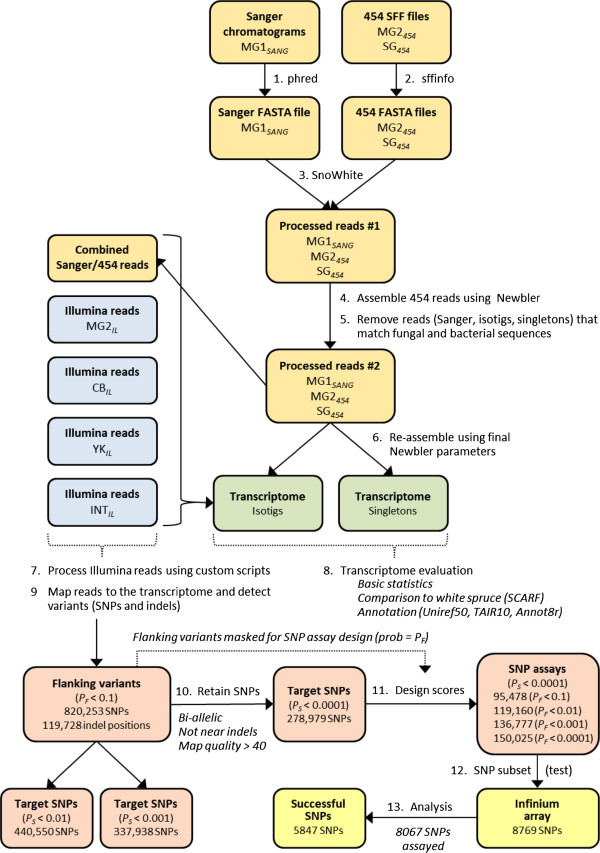
Figure 1.
Strategy for assembling the Douglas-fir reference transcriptome and detecting SNPs. We used one Sanger sequence dataset (MG1SANG) and two 454 sequence datasets (MG2454 and SG454) to assemble the reference transcriptome. We then used these same datasets plus four Illumina short read datasets (MG2IL, CBIL, YKIL, INTIL) to detect flanking variants. Orange boxes represent Sanger and 454 datasets, blue boxes represent Illumina short-read datasets, green boxes represent the reference transcriptome, red boxes represent SNP filtering steps, and yellow boxes represent SNP genotyping and analytical steps. The number of SNPs for which Infinium genotyping assays were successfully designed (Assay Design Tool score ≥ 0.6) depends on the probability used for filtering the target SNPs (PS< 0.01, 0.001, and 0.0001) and the probability used to mask nucleotides in the flanking regions (PF = 0.1, 0.01, 0.001, and 0.0001). Larger PF values resulted in more flanking variants and fewer target SNPs with successful designs.