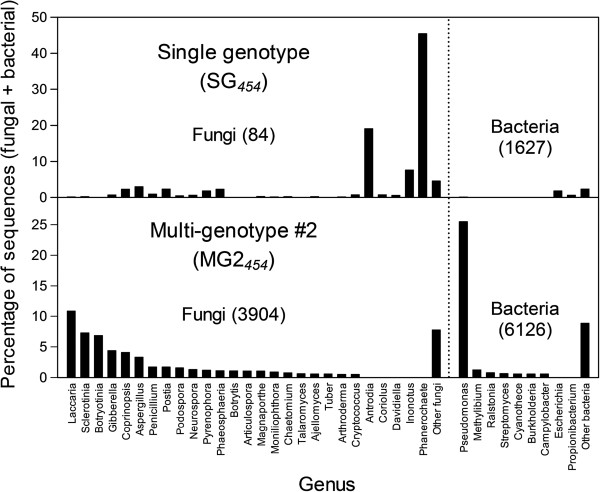
Figure 2.
Taxonomic distributions of Douglas-fir sequences identified as bacterial or fungal contaminants. We used preliminary assemblies of the SG454 and MG2454 datasets and BLAST searches to identify isotigs and singletons resulting from bacterial or fungal contamination (see Methods). Reads corresponding to these singletons and isotigs were removed prior to the final assembly. Numbers in parentheses are the total number of sequences (isotigs and singletons) in each category.