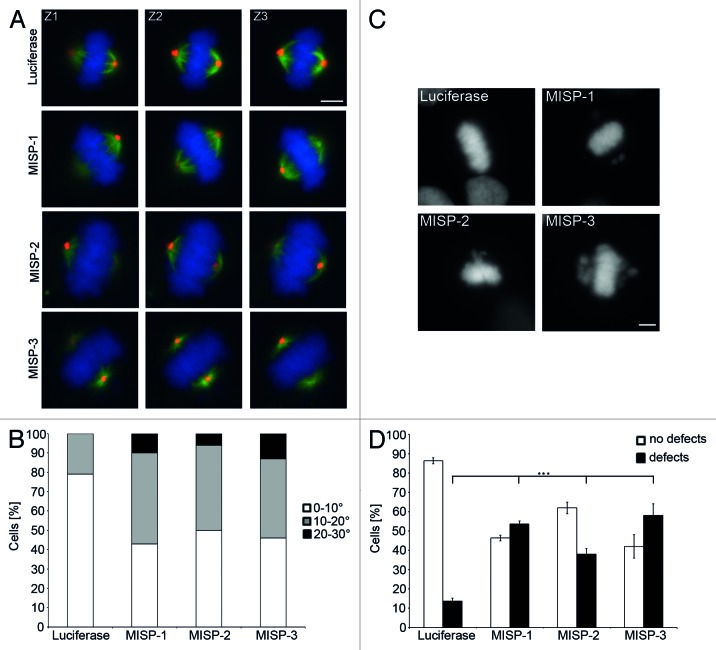
Figure 8. MISP impacts on spindle positioning and chromosome alignment. (A) Representative z-stack images (Z1, Z2, Z3 are 1 µm apart from each other) of GFP-α-tubulin-expressing UPCI:SCC114 cells immunostained with an antibody to pericentrin (red) and Hoechst 33342 (blue). Whereas centrosomes of luciferase-siRNA-transfected cells localize to the same focal plane, centrosomes in MISP-depleted cells are found in different focal planes. Cells were transfected with indicated siRNAs for 48 h. Scale bar, 10 µm. (B) The frequency distribution of the angles between substrate plane and the spindle axis is shown. The graph depicts the sum of mitotic cells evaluated in three independent experiments. Angles were determined using inverse trigonometric functions. (C) DNA staining by Hoechst 33342 of UPCI:SCC114 cells transfected with luciferase-siRNA or MISP-siRNAs for 48 h exhibits chromosomes that are not aligned at the metaphase plate in MISP-depleted cells. Scale bar, 10 µm. (D) Quantification of defects in chromosome alignment in UPCI:SCC114 cells 48 h after transfection with indicated siRNAs. The graph shows the average of three independent experiments; mean ± SD.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.