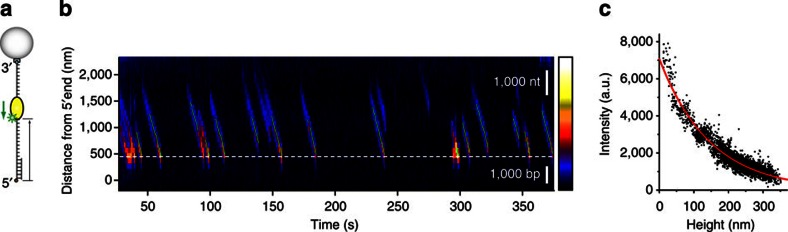
Figure 2. Imaging UvrD translocation.
(a) A partial duplex DNA construct with a long 3′-ssDNA tail. (b) A kymogram of multiple UvrD translocation events along a chosen DNA molecule stretched at 13.5 pN tension. Fluorescence intensity is colour-coded, and the colour scale is shown on the right (minimum intensity: balck (bottom); maximum intensity: white (top)). The translocation of UvrD follows a uniform motion and the speed of the translocation events are very similar, if not identical to each other. The estimated location of ssDNA/dsDNA junction is shown as the white dashed line, and the UvrD motion is observed exclusively in the ssDNA segment (above the white dashed line). Fitting the force–extension curve of this DNA molecule to the model (Supplementary Fig. S2) revealed that the length of dsDNA segment is ~1,370 bp, which is translated to 454 nm (where the dashed line is drawn) at 13.5 pN according to extensible worm-like chain model. Scale bars representing the length of 1,000 nt (ssDNA) and 1,000 bp (dsDNA) at 13.5 pN are shown in white. (c) Determination of the penetration depth of TIRF illumination. Fluorescence intensity is plotted against the height from the surface for selected translocation events at the same intensity of fluorescence excitation light (black). An exponential decay curve (red) is fit to the data to determine the penetration depth, 150 nm.