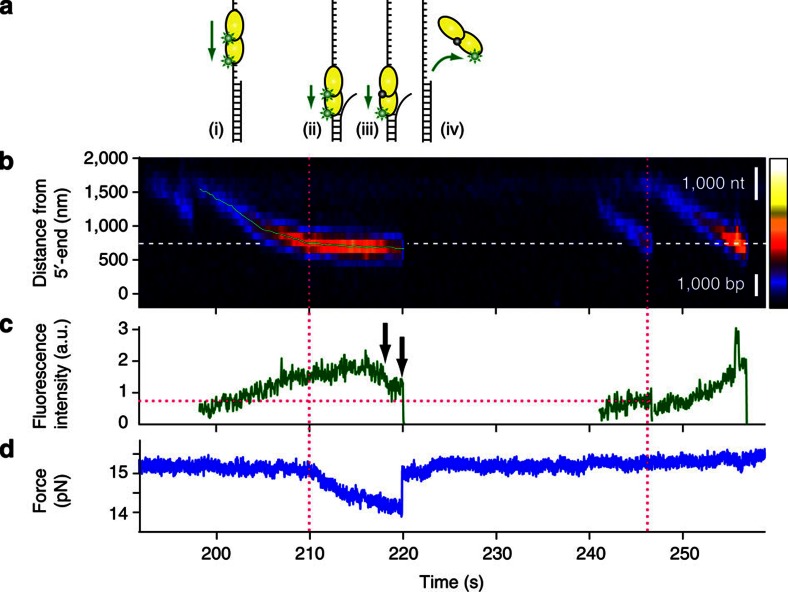
Figure 6. Two UvrD molecules translocate together and unwind the duplex segment without stalling.
(a) Schematic of this unwinding event: (i) two UvrD molecules translocate together toward the ssDNA/dsDNA junction; (ii) they proceed and unwind the junction without a pause; (iii) one fluorophore photobleaches, but the helicase keeps unwinding; and (iv) as they dissociate, the unwound duplex is immediately rezipped. (b) A kymogram of this unwinding event. White dashed line designates the estimated location of the ssDNA/dsDNA junction. Although it looks like a stalling, the position trajectory obtained from 2D Gaussian fitting (light green) reveals a slow forward motion, as expected for unwinding. For the fluorescence intensity comparison, other UvrD molecules were included. (c) The fluorescence intensity trace of the event proves that there are at least two fluorescent dyes, that is, two UvrD monomers involved in this unwinding event. First, there are two fluorescence decrease steps (black arrows). Second, the intensity of UvrD in this event is twofold higher than the intensity of another UvrD near the junction (magenta dotted lines). (d) The force trajectory. The force starts to decrease as UvrD reaches the junction (magenta dotted line). The dissociation of the enzyme from the substrate (the second black arrow) causes the duplex to rezip immediately.