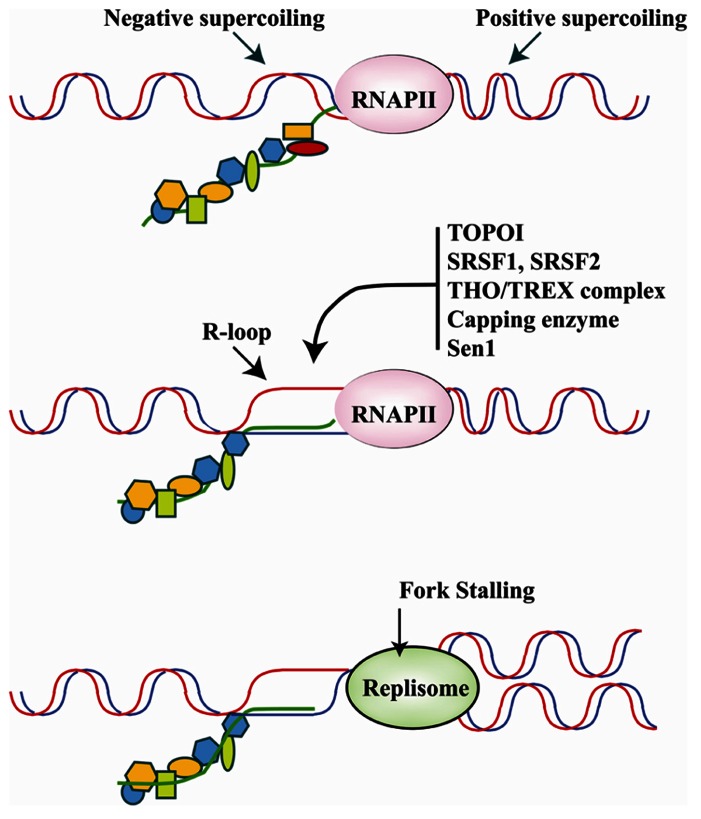
FIGURE 4.
Upper panel: the translocation of the transcriptional apparatus along the DNA induces positive and negative supercoiling, respectively, in front of and behind RNAPII. The physiological association of RBPs with the pre-mRNA molecule as it emerges from the transcriptional machinery is believed to play a major role in counteracting R-loop formation in negatively super-coiled regions. Middle panel: hybridization of the nascent RNA with the DNA template results in the formation of R-loops and occurs upon down-regulation, inhibition or mutation of several specific RBPs involved in different steps of pre-mRNA synthesis/maturation. The list of RBPs that may influence R-loop formation includes Capping enzymes, splicing factors SRSF1 and SRSF2, the THO/TREX complex, and Sen1/senataxin which is important for transcription termination. Moreover, DNA topoisomerase I (Topo I) by relieving torsional stress and phosphorylating SRSF1 can prevent R-loop formation. Bottom panel: R-loops hamper the movement of the DNA replication fork, which promotes genome instability. Thus, RBPs may be crucial to genome stability programs by inhibiting R-loop formation.