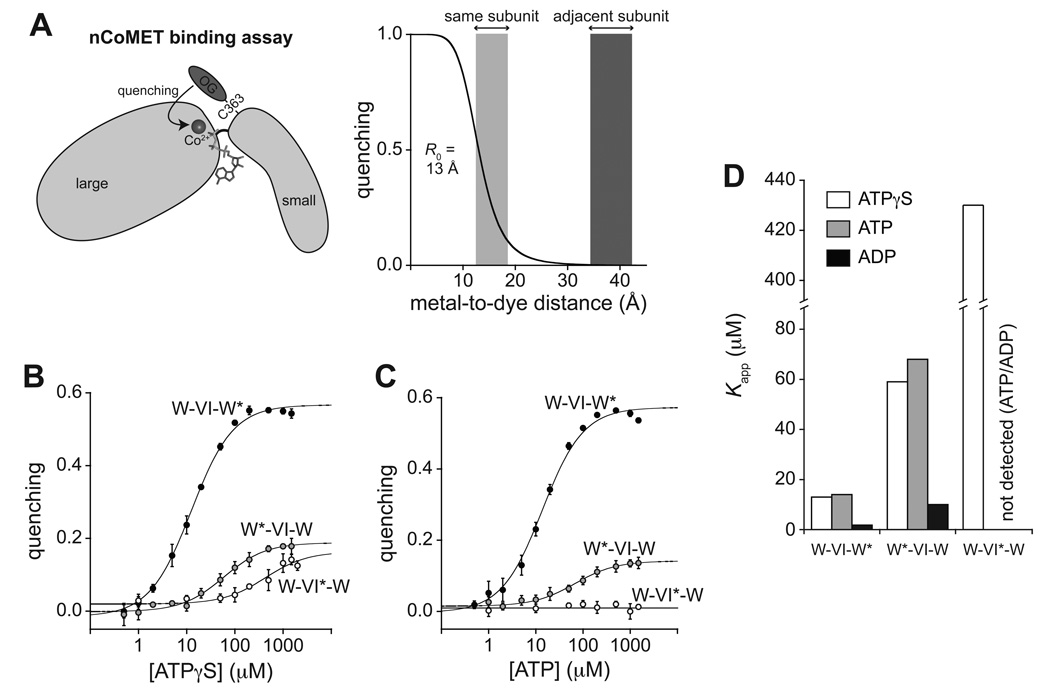
Figure 3. nCoMET detects nucleotide binding to specific subunits.
(A) In the nCoMET assay, nucleotide binds ClpX and coordinates a Co2+ ion, which quenches the fluorescence of an Oregon-Green dye attached to M363C in the small AAA+ domain of a ClpX subunit. Given the calculated R0, quenching would be substantial from nucleotide/Co2+ bound in the same subunit but minimal from nucleotide/Co2+ bound in neighboring subunits.
(B) – (C) ATPγS and ATP binding to pseudo hexamers of W*-VI-W, W-VI*-W, and W-VI-W* assayed by nCoMET. Data are shown as mean ± SD, and the lines are fits to a hyperbolic equation. Kapp values are listed in Table S2. The panel-B experiment contained 0.1 µM nCoMET variants (pseudo hexamer equivalents). The panel-C experiment contained 0.5 µM pseudo hexamer and 10 µM cp7-CFP-ssrA.
(D) Summary of fitted Kapp values for nucleotide binding to different classes of subunits. See also Fig. S3 and Table S3.