Figure 6. ATP hydrolysis by a variant with one binding site does not support function.
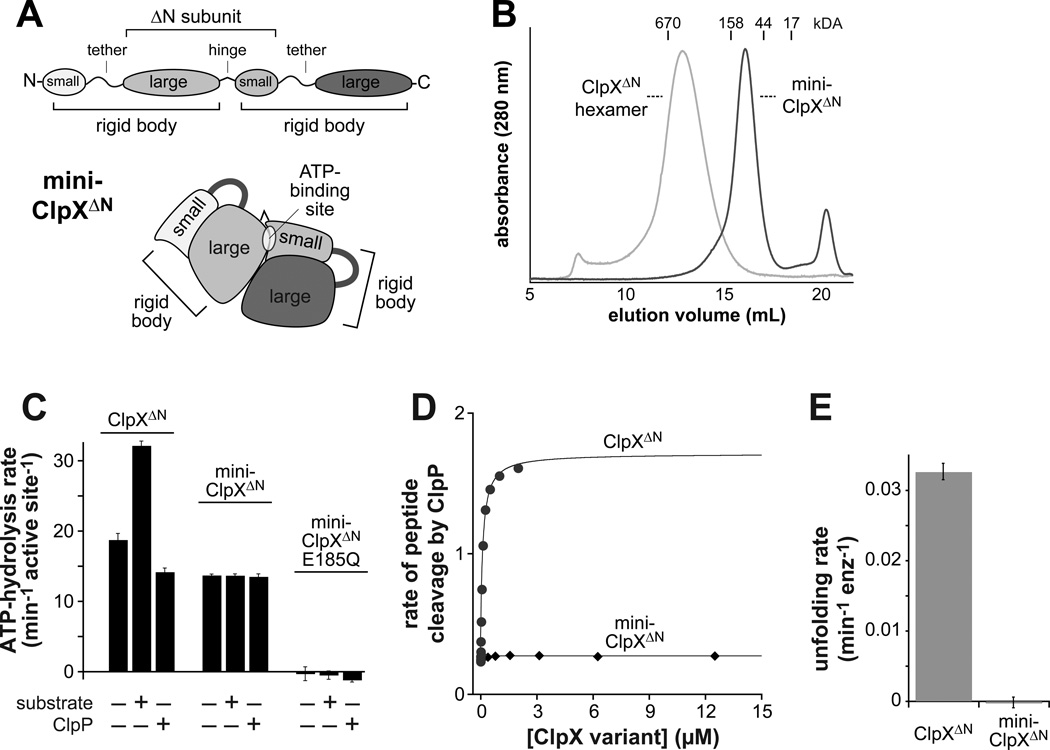
(A) Cartoon depiction of the domain structure of mini-ClpXΔN. This variant contains two rigid-body units but only one complete ATP-binding site.
(B) Mini-ClpXΔN (loading concentration 12 µM) chromatographed at a position expected for a pseudo dimer on a Superose 6 gel-filtration column. The elution position of a single-chain ClpXΔN hexamer is shown for reference.
(C) Mini-ClpXΔN hydrolyzed ATP at approximately 75% of the basal rate of single-chain ClpXΔN when activities were normalized for the number of active sites. Unlike ClpXΔN, ATP hydrolysis by mini-ClpXΔN was not stimulated by protein substrate (10 µM V15P-titinI27-ssrA) or repressed by ClpP14 (3 µM). In panels C and E, data are shown as mean ± SD.
(D) The rate of cleavage of a fluorescent decapeptide (15 µM) by ClpP14 (50 nM) in the presence of increasing concentrations of single-chain ClpXΔN or mini-ClpXΔN was determined in the presence of 1 mM ATPγS.
(E) Unfolding rates of photo-cleaved Kaede-ssrA (5 µM; Glynn et al., 2012) by single-chain ClpXΔN or mini-ClpXΔN (2 µM) were determined in the presence of 2.5 mM ATP and a regeneration system.