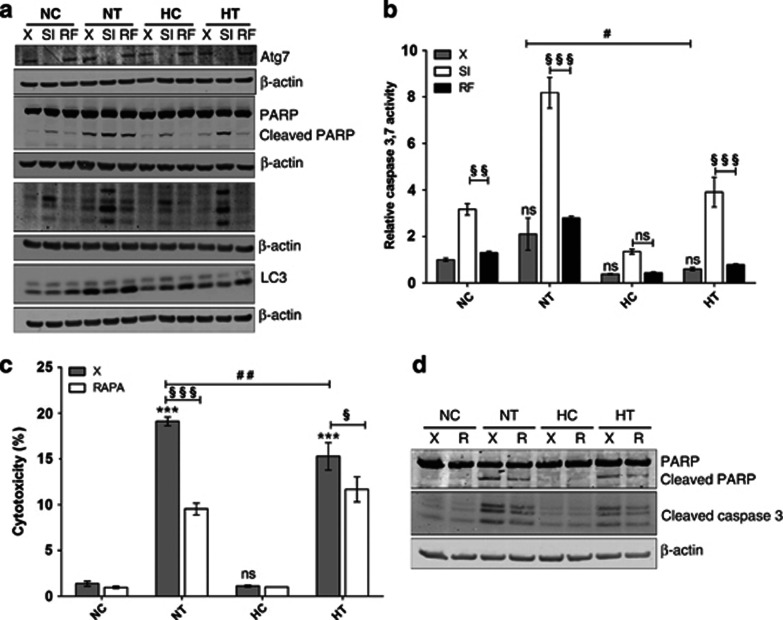
Figure 4.
Autophagy promotes cell survival after taxol incubation under normoxia and hypoxia. (a and b) MDA-MB-231 cells were untransfected (X) or transfected with Atg7 siRNA (SI) or negative control RF siRNA (RF) at 50 nM for 24 h. The transfection media were removed and replaced by culture media for 24 h. Cells were then incubated under normoxia (N) or hypoxia (H) for 16 h, without (C) or with taxol (T) at 50 μM. (a) Atg7, PARP, cleaved PARP, cleaved caspase 3 and LC3 were detected in total cell extracts by western blotting analysis, using specific antibodies. β-actin was used to assess the total amount of proteins loaded on the gel. (b) Caspase 3 and 7 activity was assayed by measuring fluorescence intensity associated to free AFC released from the cleavage of caspase 3 and 7 substrate Ac-DEVD-AFC. Results are expressed in relative caspase 3/7 activity normalized by fluorescence intensity of the control cells (NC X) and presented as means±1 S.D. (n=3). Statistical analysis was carried out with the two-way ANOVA test followed by a Bonferonni post test. NS: no significantly different from NC X; ‘#': significant difference between NT X and HT X (0.05>P>0.01); NS: no significant difference between HC SI and HC RF; ‘§§': significant difference between NC SI and NC RF (NC 0.01>P>0.001); ‘§§§': significant difference between NT SI and NT RF, HT SI and HT RF (P<0.001). (c and d) MDA-MB-231 cells were incubated under normoxia (N) or hypoxia (H), without (C) or with taxol (T) at 50 μM combined or not (X) with rapamycin (autophagy activator) at 100 nM (RAPA). (c) After 16 h of incubation, cleaved caspase 3 was detected in total cell extracts by western blotting analysis, using specific antibodies. β-actin was used to assess the total amount of proteins loaded on the gel. (d) After 40 h of incubation, LDH release was assessed. Results are expressed in percentage of viability as means±1 S.D. (n=3). Statistical analysis was carried out with the two-way ANOVA test followed by a Bonferonni post test. NS: no significantly different from NC X; ‘***': significantly different from NC X (P<0.001); ‘##': significant difference between NT X and HT X (0.01>P>0.001); ‘§': significant difference between HT X and HT RAPA (0.05>P>0.01); ‘§§§': significant difference between NT X and NT RAPA (P<0.001)