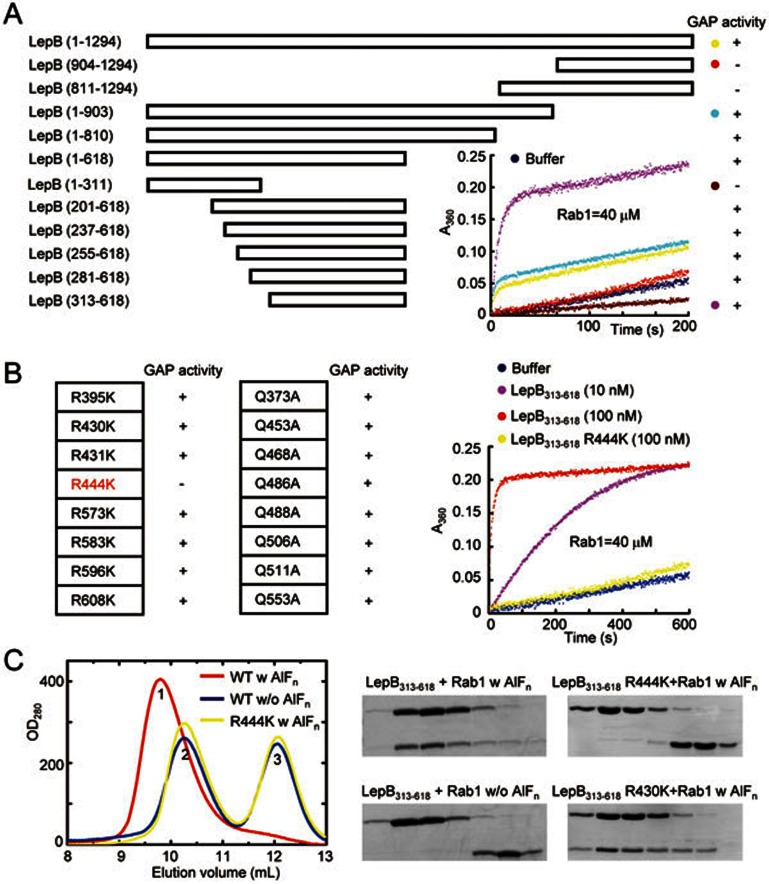
Figure 1.
Identification of the RabGAP domain of LepB and the catalytic arginine finger. (A) Mapping the RabGAP domain of LepB. A series of LepB truncation proteins as indicated were subjected to in vitro GAP assays using Rab1 as the substrate, and the positive and negative results are indicated in the right column by the + and – signs, respectively. Time courses of GTP hydrolysis for Rab1 (40 μM) catalyzed by color-indicated representative LepB truncation mutants are shown in the right corner. (B) Screening for arginine and glutamine residues required for the GAP activity of LepB313-618. Nearly all arginine and glutamine residues in LepB GAP domain (LepB313-618) were individually mutated to lysine and alanine, respectively, and each mutant protein was assayed for GAP activity using Rab1 as the substrate. The results are summarized in the left column. The time course of GTP hydrolysis for the R444K mutant in comparison with that of wild-type LepB313-618 is shown on the right. (C) Gel filtration chromatography assays for AlFn-dependent LepB-Rab1-GDP complex formation. Equal molar amounts of LepB313-618 or the R444K mutant and GDP-loaded Rab11-177 were mixed together at a total concentration of 20 mg/ml, incubated at 4 °C in a buffer supplemented with or without NaF and AlCl3, and then subjected to Superdex-75 gel filtration chromatography. Shown on the left are the chromatograms colored accordingly. Peak 1, 2, and 3 correspond to LepB313-618-Rab1-GDP-AlFn complex, free LepB313-618, and free Rab1, respectively. Elution fractions in the indicated range were analyzed by SDS-PAGE and Coomassie blue-stained gels are shown on the right. LepB313-618 R430K was included as a negative control.