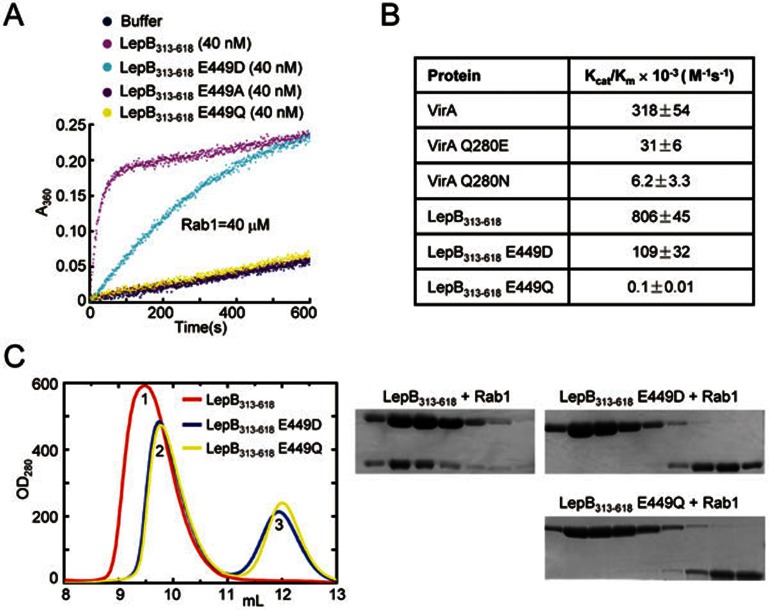
Figure 4.
Glu449 is required for the RabGAP activity of LepB as well as AlFn-mediated LepB-Rab1-GDP complex formation. (A) Effects of Glu449 mutations on the RabGAP activity of LepB. Time courses of GTP hydrolysis for Rab1 (40 μM) catalyzed by LepB313-618 or its E449A, E449D and E449Q mutants are shown as color indicates. (B) Summary of the kinetic parameters of LepB Glu449 and VirA Gln280 mutants. The Michaelis-Menten kinetic parameters (kcat/Km) determined by using the Lineweaver-Burk plot for LepB313-618 and VirA or their indicated mutants are listed. Data shown are mean values ± standard deviation (error bar) from two independent experiments. (C) Effects of Glu449 mutations on AlFn-mediated LepB-Rab1-GDP complex formation. The assay was performed similarly as that in Figure 1C. Shown on the left are the gel filtration chromatograms colored accordingly. Peek 1, 2, and 3 correspond to LepB313-618-Rab1-GDP-AlFn complex, free LepB313-618, and free Rab1, respectively. Elution fractions in the indicated range were analyzed by SDS-PAGE and Coomassie blue-stained gels are shown on the right.