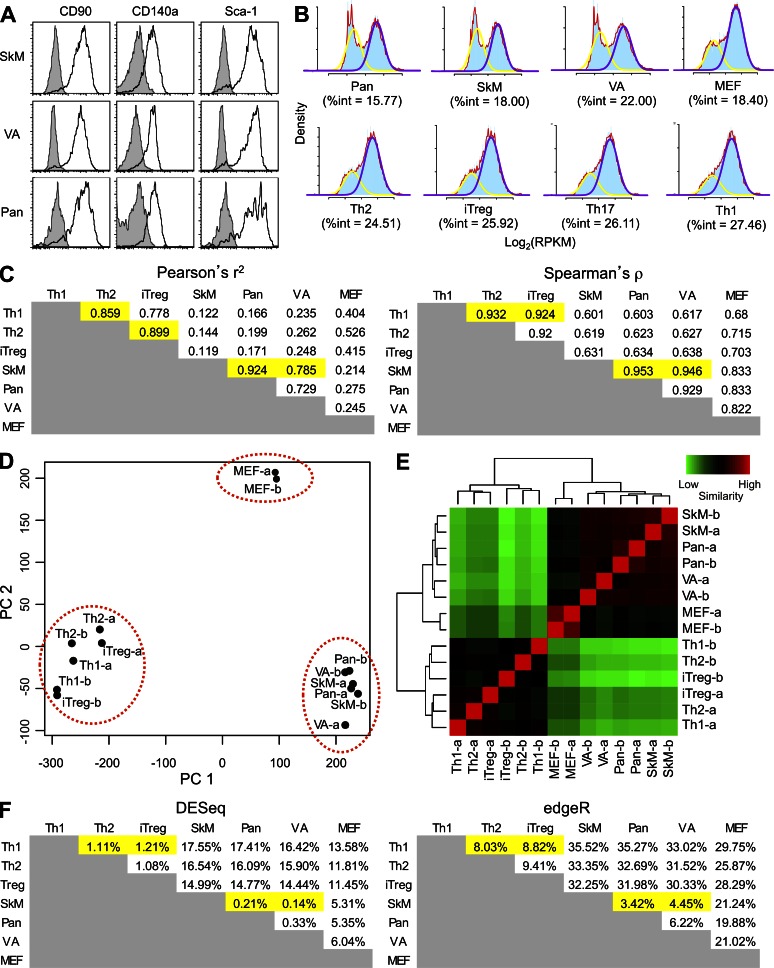
Figure 2.
Analysis of the transcriptomes of FAP+ cells from adipose tissue, skeletal muscle, and pancreas. (A) FAP+ cells were assessed by flow cytometry for their expression of the membrane proteins CD90 (Thy1), CD140a (PDGFRα), and Sca-1 (Ly6A/E), which are characteristic of certain mesenchymal cells. Filled histograms represent isotype staining, and the black lines denote the specific stains. (B) The RNA-Seq data for each of the cell types were log transformed and modeled as bimodal distributions. The %int represents the degree of heterogeneity within each population. The density value is indicated in blue. The yellow line represents the bimodal fit for the lowly expressed transcripts and the purple line for the highly expressed transcripts, and the red line indicates the overall density value. (C) The Pearson’s r2 and Spearman’s ρ correlations were calculated for the RPKM gene expression levels of FAP+ cells, FAP− MEFs, and CD4+ T cell subsets after removal of low expressed/nonexpressed genes. (D) PCA analysis of VST (counts) of FAP+ cells, CD4+ T cell subsets, and MEFs is shown. (E) VST clustering of RPKM levels of replicate samples of FAP+ cells, CD4+ T cell subsets, and MEFs is shown. (F) The percentage of differentially expressed genes among the different cells was determined by DESeq and edgeR using count data. The highlighted cells here and in C show the correspondence between high levels of similarity in these two analyses of gene expression. Th1, Th2, and iTreg refer to the CD4+ T cell subsets. VA, SkM, and Pan refer to FAP+ cells from visceral adipose tissue, skeletal muscle, and pancreas, respectively. See also Table S1.