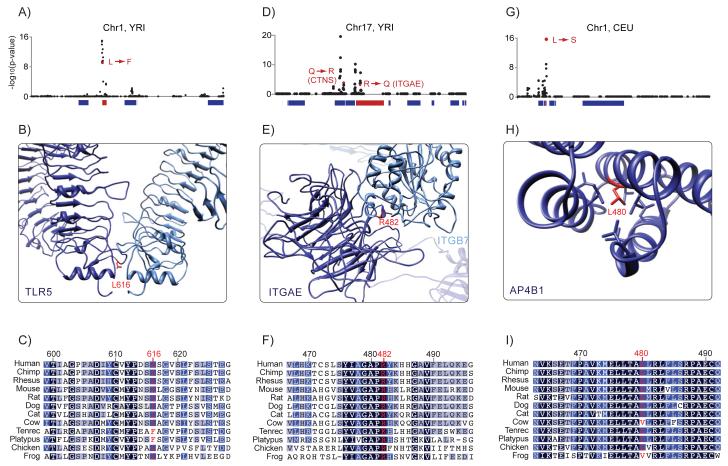
Figure 2. Candidate Non-Synonymous Mutations Identified by CMS.
CMS identified high-scoring non-synonymous mutations in the genes (A-C) TLR5, (D-F) ITGAE and (G-I) AP4B1. Panels (A), (D), and (G) show CMS scores for all variants in the regions. High-scoring non-synonymous variants and the genes in which they are located are presented in red. Panels (B), (E), and (H) show homology modeling of the genes with the residue containing the candidate variants in red. Panels (C), (F), and (I) show the amino-acid sequence in 12 vertebrate species. The color of the residue indicates the conservation score (darker color indicates greater conservation). See also Table S6.