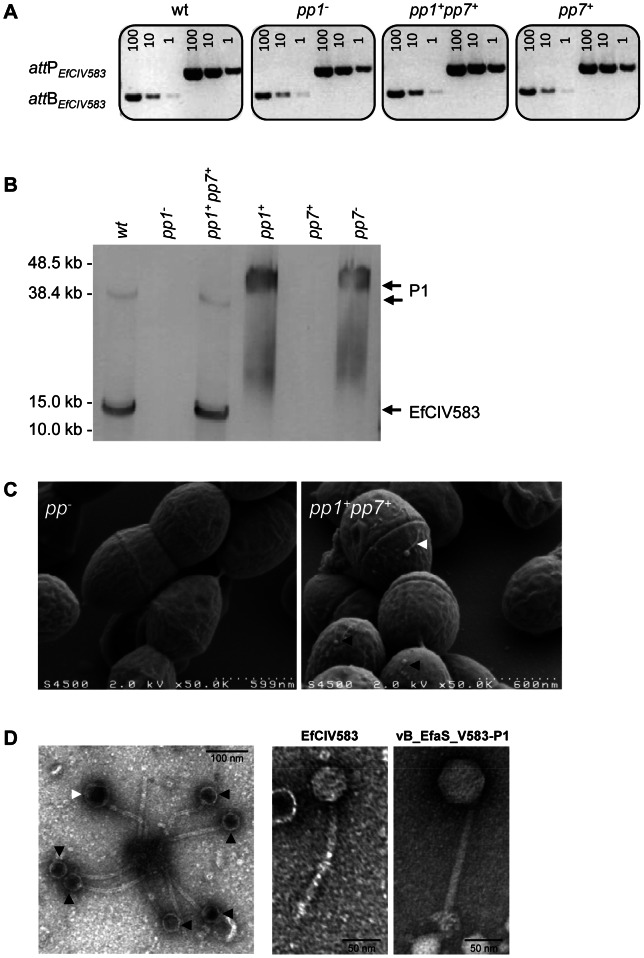
Figure 4. Interaction between E. faecalis pp1 and pp7 (EfCIV583).
(A) Semi-quantitative PCR detection of EfCIV583 circular forms (attP) and excision sites (attB) in wild-type (WT) and strains pp1−, pp1+ pp7+ and pp7+. Excision and circularization products probed by semi-quantitative PCR on 100, 10 and 1 pg of total bacterial DNA prepared from cultures of WT and strains pp1−, pp1+ pp7+ and pp7+ induced for 2 h with 2 µg/ml of ciprofloxacin at 37°C. Twenty cycles were used to amplify products of pp1 and EfCIV583. These results are representative of two independent experiments. (B) Prophage DNA extracted from precipitated phage particles obtained from lysates of WT and strains pp1−, pp1 + pp7 + and pp7 + was separated by FIGE and analyzed by Southern-blot and hybridized sequentially using specific probes for pp1 and EfCIV583 genomes. The approximately 38.2 kb and 12 kb band corresponds to P1 and EfCIV583 genome, respectively. As ascertained by pp1-specific hybridization, migration of P1 DNA was delayed in lane pp1 + and pp7 − compared to lanes WT and pp1 + pp7 +. Lambda DNA mono-cut mix (NEB) was run next to the samples to validate band sizes. (C) Scanning electron microscopy images of bacterial cells from strains pp− and pp1 + pp7 + after ciprofloxacin treatment. (D) Transmission electron microscopy images of phages produced by strain pp1 + pp7 + after ciprofloxacin treatment. White and black arrows indicate big and small sized particles attributed to P1 and EfCIV583, respectively. Enlarged images of EfCIV583 and P1 (renamed vB_EfaS_V583-P1) are shown on the right.