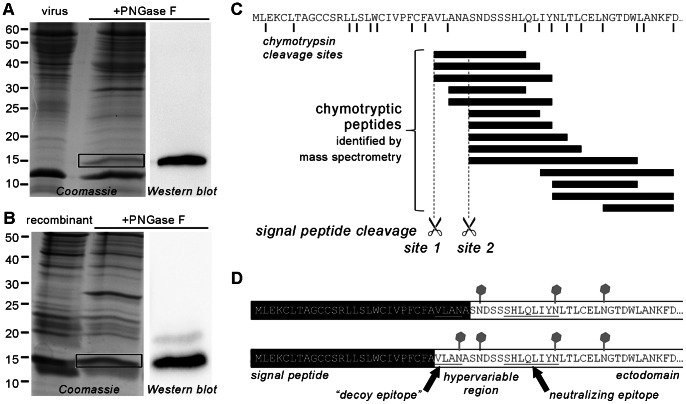
Figure 7. Identification of the signal peptide cleavage site of GP5 (virus-derived and recombinant) by mass spectrometry.
(A), PRRSV (strain VR-2332) was grown in MARC-145 cells, precipitated with PEG-8000, pelleted and subjected to sucrose density gradient centrifugation. The virus-containing fraction was left untreated or deglycosylated with PNGase F and separated by reducing SDS-PAGE followed by Coomassie staining (left-hand side) or Western blot (anti-GP5 antiserum, right-hand side). The deglycosylated band corresponding to GP5 (black box) was cut out of the gel, digested with trypsin or chymotrypsin and analyzed by LC-MS/MS. (B), PRRSV GP5 (with His tag) and M (with HA tag) were co-expressed in Sf9 insect cells by infection with recombinant baculovirus. Following cell harvesting and lysis, GP5–M was enriched using Ni-NTA agarose (binding to GP5–His). The eluated protein was left untreated or digested with PNGase F and subjected to reducing SDS-PAGE and Coomassie staining (left-hand side) or Western blot (anti-His-tag antibody, right-hand side). The deglycosylated GP5 band (black box; coinciding with M) was cut out and treated as in (A). (C), representative result from mass spectrometry of virus-derived GP5 (as in A). The first 61 residues of GP5 are shown with the positions of the predicted chymotrypsin cleavage sites (black lines) and the putative signal peptide cleavage sites (broken lines). Chymotryptic peptides that were identified are represented as black bars. The pattern of peptides is evidence for signal peptide cleavage at sites 1 and 2. No peptides corresponding to the signal peptide region (1–26) were identified. (D), Conclusion from mass spectrometry, showing the N-terminal sequence of GP5 with signal peptide (black), glycosylations (grey) and the positions of the neutralizing and the “decoy epitope”. Two GP5 species exist with signal peptide cleavage at sites 2 (top) and 1 (bottom), respectively.