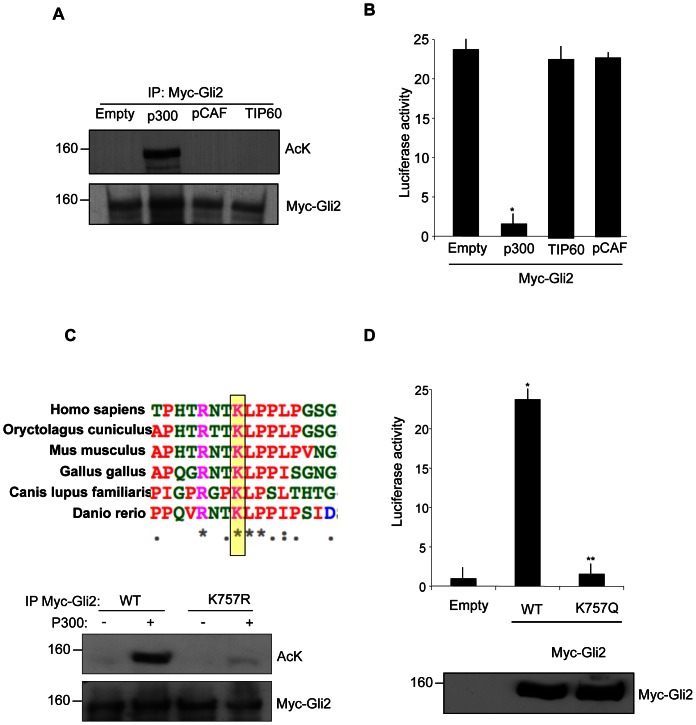
Figure 1. p300 acetylates Gli2 at the conserved Lysine 757.
(A) In vivo acetylation assay in HEK293T cells transfected with plasmids coding for Myc tagged Gli2 and the indicated HATs. Cell extracts were immunoprecipitated with anti-Myc antibody and acetylation was revealed with anti-acetyl lysine antisera. Western blot analysis with anti Myc antibody showed equivalent amounts of Myc-Gli2. Empty, pcDNA3. (B) Luciferase assay in HEK293T. Cells were transfected with 12× Gli-Luc and TK renilla reporter vectors, Gli2, p300, TIP60, pCAF where indicated. *p<0.01 vs Empty (pcDNA3). Results are shown as the average ± SD of triplicate experiments (n = 4). (C) (Top) ClustalW alignment of the region surrounding lysine 757 of Gli2 in different species. Identical, strongly conservative and weakly conservative aminoacids are indicated by asterisk, colon and dots respectively, according to ClustalW convention. (Bottom) In vivo acetylation assay in HEK293T cells transfected with Myc-Gli2 WT or K757R and p300 where indicated. Acetylation was detected as previously described. (D) (Top) Luciferase assay performed in HEK293T cells transfected with 12× Gli-Luc, TK renilla, Myc-Gli2 WT or the acetylation mimetic mutant Myc-Gli2 K757Q. *p<0.01 vs Empty; **p<0.05 vs WT. Results are shown as the average ± SD of triplicate experiments (n = 3). (Bottom) Western blot analysis of Myc-Gli2 WT and K757Q expression levels.