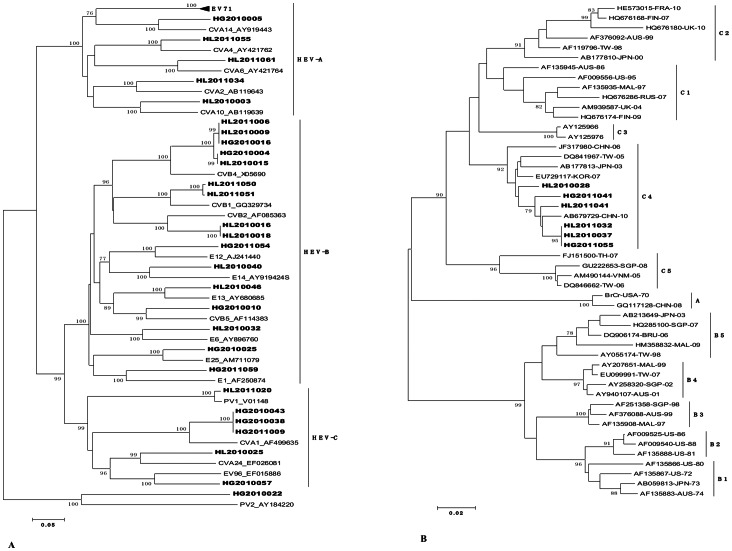
Figure 2. A. Rooted phylogenetic trees of partial VP1 sequences from clinical specimens collected from healthy children with EV infection from 2010 to 2011 and reference sequences available in GenBank for each serotype.
The evolutionary distances were calculated using the Kimura two-parameter model for nucleotide substitution and the neighbor joining method to reconstruct the phylogenetic tree (MEGA version 5.0). Sequence names for field strains are constructed as follows: municipality number (starting with HL for the Longgang residential district of Shenzhen and with HG for the Guangming residential district of Shenzhen). The 21 reference sequences have GenBank accession numbers, whereas sequences generated in the present study are in bold letters. B. Phylogenetic tree depicting the relationships among the VP1 sequences of the EV71 isolates.