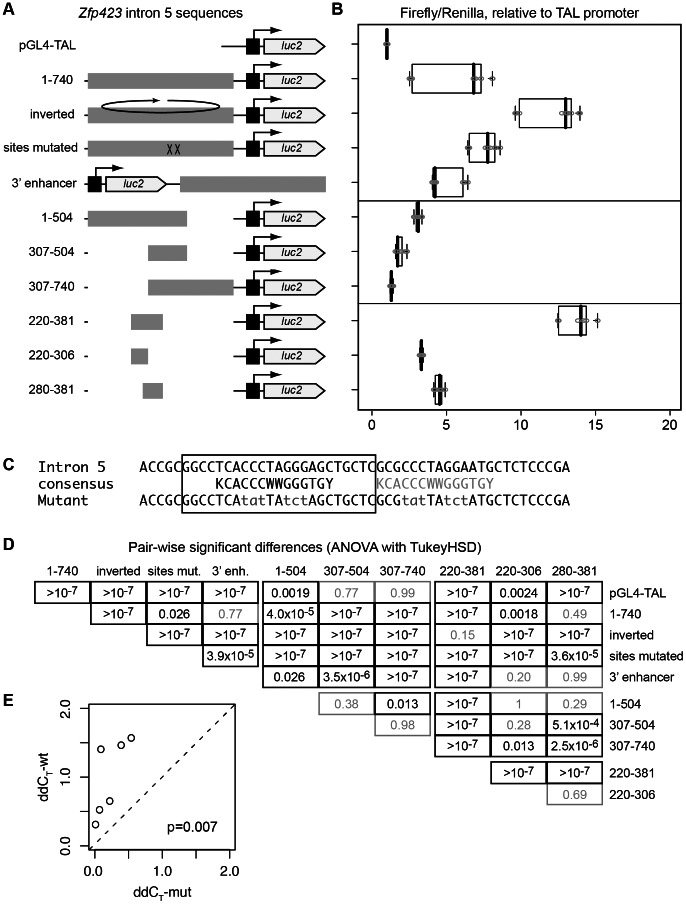
Figure 4. Zfp423 intron 5 binding site is an enhancer.
(A) Reporter constructs are shown schematically. A pTAL minimal promoter was inserted upstream of the firefly luciferase reporter in pGL4. Fragments of the Zfp423 intron 5 putative enhancer were placed as indicated by the alignment of grey boxes. Position of the Zfp423 consensus match is indicated by XX in the “sites mutated” construct. Horizontal lines indicated constructs tested together, with pGL4-TAL repeated in each group. (B) Reporter gene expression levels, expressed as the ratio of firefly luciferase to co-transfected Renilla luciferase enzymatic activity measured by luminescence. Each measurement was normalized to the average of control vector (pGL4-TAL) samples run on the same day. For each construct, 2–3 DNA preparations were each used in 2–4 independent co-transfections for 9–12 (top eight constructs) or 6 (bottom three constructs) measurements. (C) Sequence changes in the sites mutated construct. Top line shows sequence of the mouse reference clone. The overlapping ROAZ (Zfp423), OLF1 site predicted by SynoR is boxed. Consensus Zfp423 (ROAZ) binding site and an adjoining consensus half-site (grey text) are indicated. Bottom line shows mutated sequence, with altered residues in grey lowercase. (D) As one estimate of the significance level for apparent shifts between constructs, normalized expression values were compared by ANOVA, followed by Tukey's Honest Significant Differences pair-wise comparisons. Calculated p-values are indicated in the intersection between construct designations. Potential scaling differences between sequential experiments on different days (indicated by gaps between cells in the table) may violate some assumptions of the test. (E) ChIP-qPCR performed for transfected plasmids shows a higher enrichment index compared to input and IgG controls (calculated as ΔΔCT for carrying the wild-type site than for the mutated sites shown in (C). The pairwise comparison was significant at p = 0.007 by t-test or 0.03 by the nonparametric equivalent (Wilcoxon signed rank test).