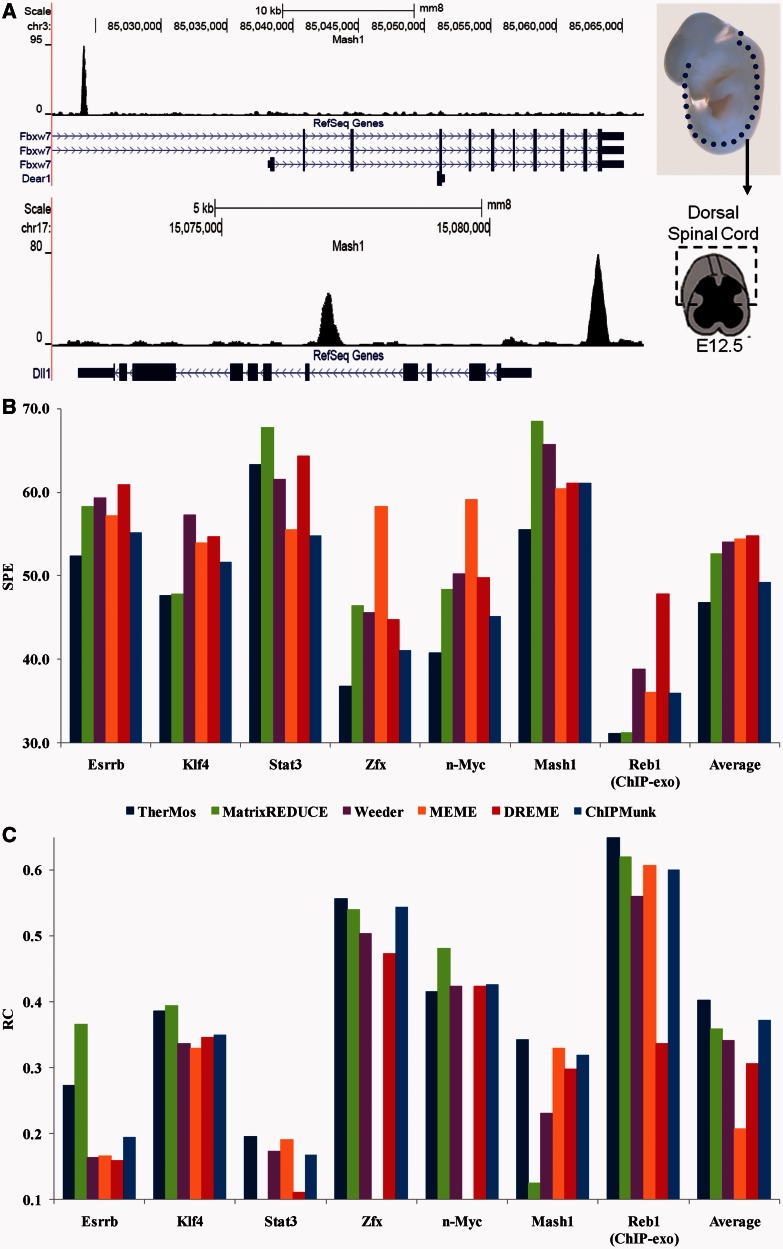
Figure 2.
(A) Mash1 in vivo ChIP-seq profile (E12.5 mouse spinal cord) shows strong peaks at known targets of Mash1. (B, C) Performance of TherMos and other algorithms in 10-fold cross-validation testing on the seven whole-genome TF binding profiles. For each algorithm and each TF, the bar height indicates the average SPE or rank correlation coefficient across the 10 test sets. The summary bars at the end indicate average performance across all seven TFs. (B) SPE is calculated between predicted (motif) and observed (experimental data) ChIP-seq binding profile. Smaller SPE indicates higher accuracy. (C) Rank correlation coefficient is calculated between predicted (motif) and observed (experimental data) ChIP-seq tag counts. Average rank correlation coefficients below zero for some of the algorithms are not shown.