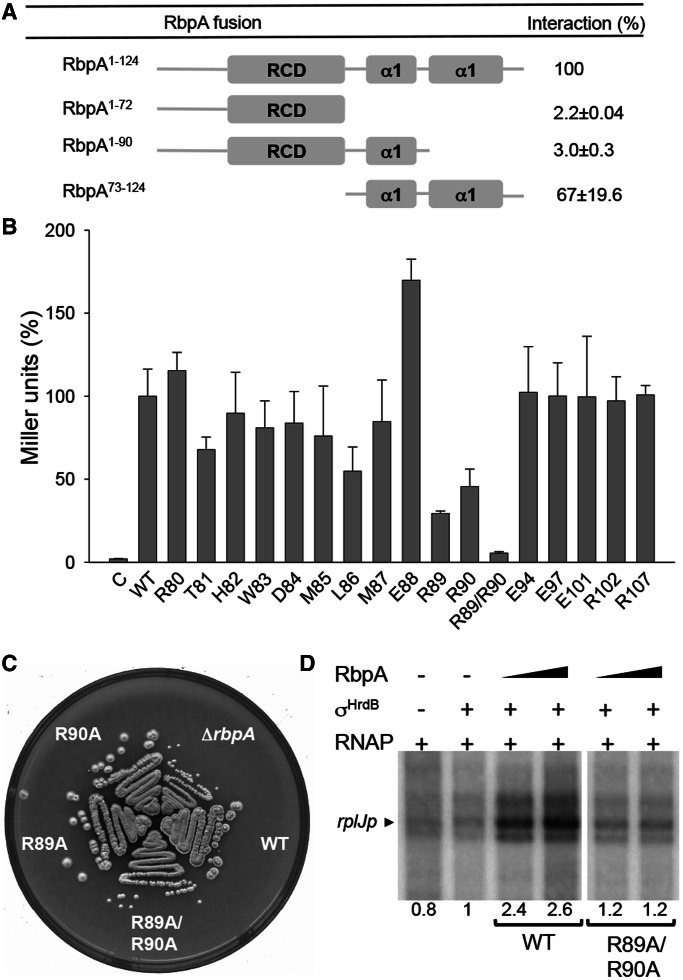
Figure 6.
The C-terminal region of RbpASc is necessary and sufficient for σHrdB interaction. (A) A schematic diagram indicating the regions of RbpASc tested for interaction with σHrdB using BACTH analysis. The RbpASc fragments and σHrdB σ2–σ4 were fused to the T18 and T25 domains of B. pertussis adenylate cyclase, respectively. (B) BACTH analysis of alanine point mutations in conserved residues in the C-terminal region of RbpASc. C, control strain containing pKT25–hrdB and pUT18. For (A) and (B), experiments were performed in triplicate (standard deviations indicated), and data are presented as % Miller units relative to results obtained with the full-length wild-type RbpASc. Control strains with pKT25–hrdB and pUT18 exhibited <1% the activity of the full-length RbpASc interaction. (C) Growth of S129 (ΔrbpA), S129 containing pSETΩ::rbpA (WT), or equivalent constructs with the rbpA mutations as indicated. Strains were streaked to MS agar and photographed after 4 days incubation at 30°C. (D) In vitro transcription from the rplJ promoter in the presence of RbpASc or RbpASc R89A/R90A mutant proteins. Multi-round in vitro transcription reactions contained core RNAP (50 nM) σHrdB (250 nM), RbpASc (250 nM or 1.25 µM) and a DNA template generated by PCR using primers listed in Supplementary Table S2. Transcript levels were quantified by phosphorimaging, and data are presented relative to reactions performed in the presence of σHrdB but the absence of RbpA.