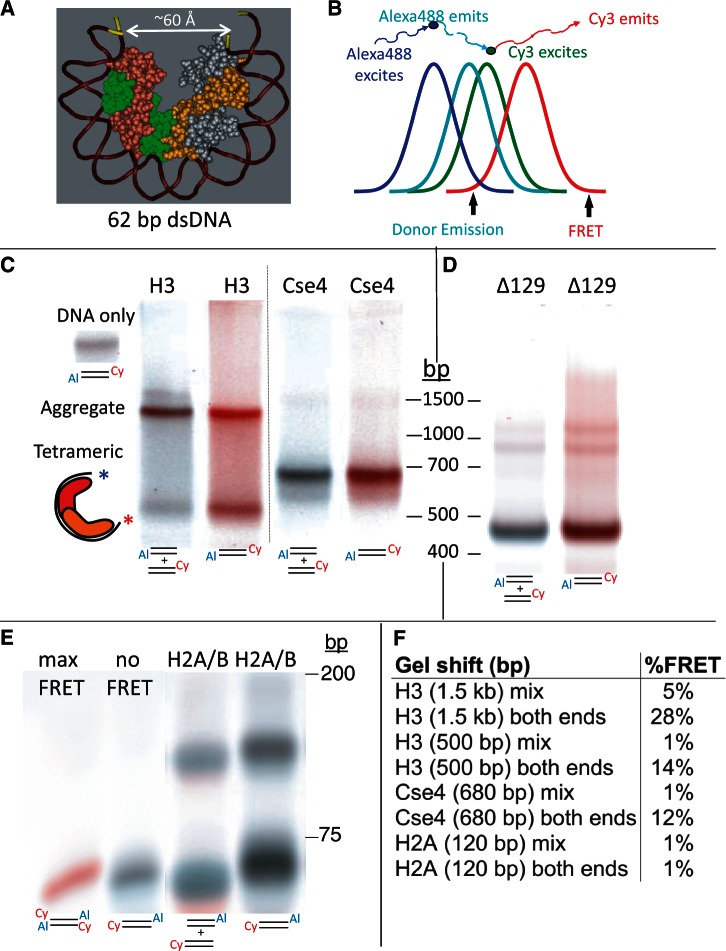
Figure 4.
A gelFRET assay directly confirms hemisome formation. (A) Structural model based on PDB 1KX5, showing that a 62-bp duplex is sufficient to wrap a hemisome. (B) FRET strategy using Alexa488 as donor and Cy3 as acceptor, where colors illustrate wavelength and curves represent approximate spectral overlap. (C) GelFRET analysis of a representative gel-shift experiment in which H3 or Cse4 octamers and mixed singly or doubly end-labeled duplexes (as indicated below each lane with 5′ Cy3 on α62 R and 3′ Alexa488 on α62 F) were combined in 2 M NaCl, subjected to dialysis versus 10 mM HEPES (pH 7.5) + 0.25 mM EDTA and electrophoresed on a native 6% polyacrylamide gel, followed by scanning for gelFRET. A doubly end-labeled DNA band showing no FRET signal served as a negative control. Al = Alexa 488. Cy = Cy3 (D) Deletion of the Cse4 N-terminal tail increases gel-shift mobility. Octamers containing tail-deleted Cse4 were assembled with α62 DNA duplexes labeled for gelFRET. DNA standards are included only as a rough guide. (E) As a negative control, H2A/H2B dimers were used for assembly (Supplementary Figure S5). When DNA fragments are in large molar excess, single H2A/H2B dimers bind and cause a gel shift (34). Rapidly migrating dye-coupled fragments resolve from one another with Alexa488 (which is coupled via an amino-C7 linker) causing slower migration than Cy3 (which is directly coupled) of otherwise identical fragments, and resulting in misalignment of mixed Cy3 and Alexa488 fragments. (F) Quantification of gelFRET emissions for the bands shown in the figure.