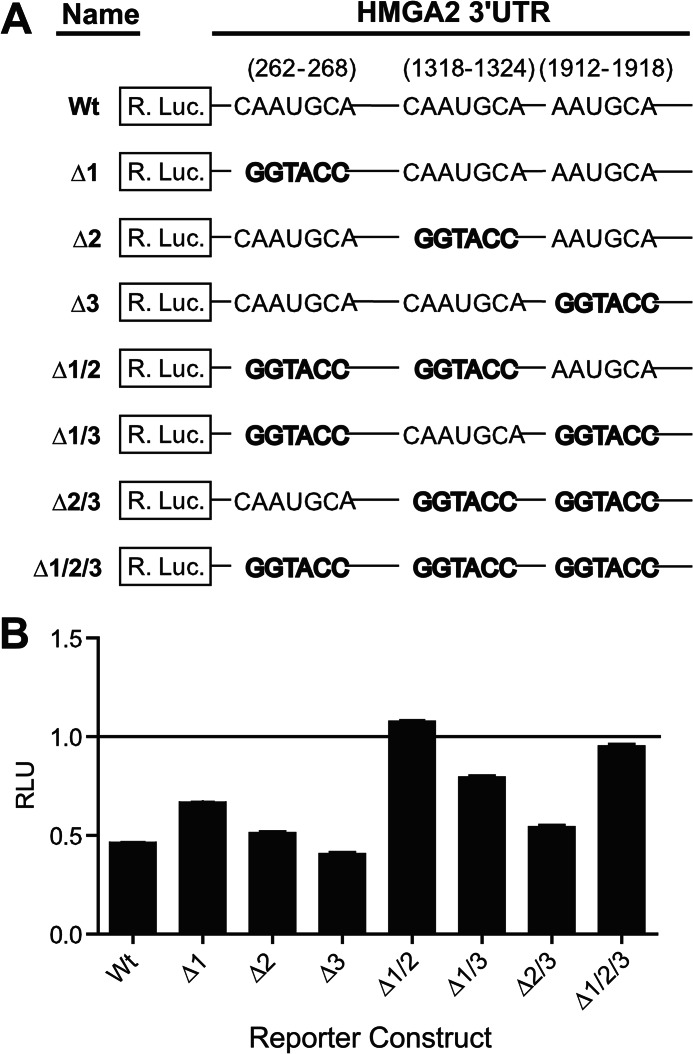
FIGURE 7.
Characterization of the three predicted miR-33a binding sites located within the HMGA2 3′-UTR. A, diagram depicting the HMGA2 3′-UTR reporter construct mutants used to characterize functional miR-33a sites. The miR-33a seed sequences were mutated by replacement with a KpnI restriction enzyme recognition sequence (GGTACC). B, NCI-H1299 cells, transfected with individual HMGA2 3′-UTR reporter constructs from A and an miR-33a mimetic oligonucleotide or a scrambled control oligonucleotide (each at 20 nm), were assayed for luciferase activities 48 h post-transfection. Relative luminescence units (RLU) were normalized to the corresponding scrambled control for each reporter construct (n = 3). Error bars, S.D.