FIGURE 4.
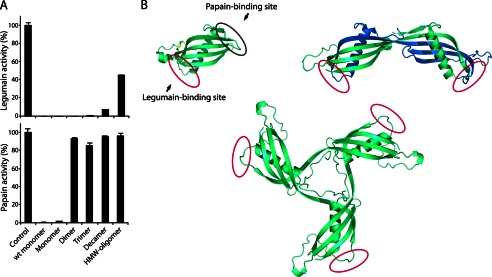
Oligomers are domain-swapped and retain a native-like fold. A, inhibitory capacity of isolated cystatin C oligomers tested against the two proteases, legumain and papain, both of which are inhibited by monomeric cystatin C. The control samples represent the full protease activity without inhibitors. Native, nonstabilized, monomeric cystatin C is denoted wt monomer. Monomer, Dimer etc. refer to disulfide-stabilized molecules. Error bars, S.D. B, crystal structure of monomeric stab1 cystatin C (Protein Data Bank entry 3GAX) (25) displaying the two protease binding sites residing on opposite sides of the molecule, where the N-terminal and two hairpin loops (L1 and L2) constitute the papain-binding domain and the legumain-binding domain resides in proximity to Asn39 (20). The crystal structure of dimeric cystatin C (Protein Data Bank entry 1TIJ) shows disruption of the papain-binding loop L1 that acts as a hinge to the swapping domain, whereas the fold of the legumain-binding domain is retained and accessible. The three-dimensional model of trimeric cystatin C was constructed in PyMol by repositioning the domain swapping structural elements (residues 1–57) in the monomer (Protein Data Bank entry 1TIJ) (44). The oligomer model is in agreement with previous experimental data obtained for dimeric wt cystatin C and the presented inhibitory profiles for stabilized oligomeric cystatin C.
