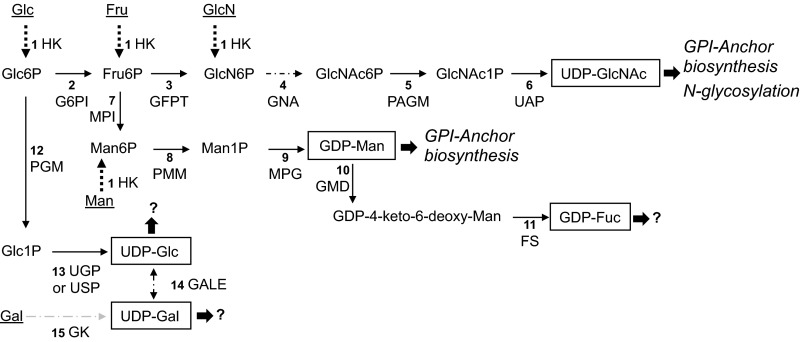
FIGURE 1.
Sugar nucleotide biosynthetic pathways identified in the genome of P. falciparum. The numbers refer to the enzymes and known or candidate genes described in Table 1. Predicted known fates of sugar nucleotide donors according to the glycoconjugates described in P. falciparum are in italic type (or marked with a question mark if the fate is unknown). Sugar nucleotides identified in this study are boxed. Dotted lines indicate confirmed salvage pathways, and sugars, taken up from the medium, are underlined. The discontinuous arrow (step 4) represents the glucosamine-phosphate N-acetyltransferase activity (EC 2.3.1.4) for which a candidate gene is not yet identified. HK, hexokinase; G6PI, glucose-6-phosphate isomerase; GFPT, glucosamine-fructose-6-phosphate aminotransferase; GNA, glucosamine-phosphate N-acetyltransferase; PAGM, phosphoacetylglucosamine mutase; UAP, UDP-N-acetylglucosamine pyrophosphorylase; MPI, mannose-1-phosphate isomerase; PMM, phosphomannomutase; MPG, mannose-1-phosphate guanyltransferase; GMD, GDP-mannose 4,6-dehydratase; FS, GDP-l-fucose synthase; PGM, phosphoglucomutase; UGP, UTP-glucose-1-phosphate uridylyltransferase; GALE, UDP-glucose 4-epimerase; GK, galactokinase. The suggested pathway for the biosynthesis of UDP-Gal through the activity of the UDP-sugar pyrophosphorylase enzyme is indicated with a gray discontinuous arrow (see “Discussion”).