Background: pVHL, a tumor suppressor, functions as the substrate recognition component of an E3-ligase complex that targets hypoxia inducible factor (HIF) 1α for destruction.
Results: pVHL binds ribosomal protein RPS3, interferes with ribosome assembly, and inhibits protein synthesis.
Conclusion: pVHL suppresses ribosome biogenesis and protein synthesis.
Significance: The findings disclose a novel function of pVHL and provide insight into the regulation of ribosome biogenesis.
Keywords: Cancer Biology, Protein Synthesis, Ribosome Assembly, Ribosomes, Tumor, pVHL
Abstract
pVHL, the product of von Hippel-Lindau (VHL) tumor suppressor gene, functions as the substrate recognition component of an E3-ubiquitin ligase complex that targets hypoxia inducible factor α (HIF-α) for ubiquitination and degradation. Besides HIF-α, pVHL also interacts with other proteins and has multiple functions. Here, we report that pVHL inhibits ribosome biogenesis and protein synthesis. We find that pVHL associates with the 40S ribosomal protein S3 (RPS3) but does not target it for destruction. Rather, the pVHL-RPS3 association interferes with the interaction between RPS3 and RPS2. Expression of pVHL also leads to nuclear retention of pre-40S ribosomal subunits, diminishing polysomes and 18S rRNA levels. We also demonstrate that pVHL suppresses both cap-dependent and cap-independent protein synthesis. Our findings unravel a novel function of pVHL and provide insight into the regulation of ribosome biogenesis by the tumor suppressor pVHL.
Introduction
Protein synthesis is a fundamental cell process with ribosomes assigned the task of decoding the genetic code. Ribosome biogenesis is exceptionally complex and in eukaryotic cells occurs mainly in the nucleolus requiring a myriad of proteins. The pathway begins with transcription of ribosomal proteins and ribosomal RNA (rRNA) precursors. The association of ribosomal proteins and non-ribosomal factors with nascent pre-rRNAs gives birth to a 90S pre-ribosomal complex. The 90S complex separates into a pre-60S complex that will yield the large ribosomal subunit and a pre-40S complex that will generate the small ribosomal subunit (1–3).
Ribosome production is enhanced in cancer cells and associated with profound quantitative and qualitative alterations in ribosomal biogenesis and function (4). Indeed, the synthesis of rRNAs, as well as the production of several ribosomal proteins, is up-regulated in a variety of tumors (5). As well, several tumor suppressors and oncogenes influence the formation of ribosomes. For example, loss of two major tumor suppressor gene products, the retinoblastoma protein (pRB) and p53, stimulates ribosome biogenesis in cancer cells (6). pRB is a suppressor of rRNA synthesis (7, 8) and p53 inhibits ribosome biogenesis through repressing the transcription of both RNA polymerase I and III (9). Notably, phosphatase and tensin homolog deleted in chromosome 10 (PTEN) also represses polymerase I transcription and MYC increases polymerase I activity and transcription of polymerase II and III genes (10). However, mechanistic links between these aforementioned events and ribosome assembly is lacking. Eukaryotic ribosome assembly is dependent on the presence of a large number of rRNA precursors and ∼190 proteins have been implicated in eukaryotic ribosome biogenesis, stemming from the initial nucleolar steps to final processing in the cytoplasm (11, 12). These pre-ribosomal factors are, for the most part, essential for ribosome biogenesis (11).
Silencing or mutation of the von Hippel-Lindau (VHL)2 tumor suppressor gene leads to a variety of tumors, including the clear cell renal carcinoma, pheochromocytoma, hemangioblastoma, and polycythemia (13, 14). pVHL, the product of VHL, is an adaptor protein implicated in many biological processes (15). For example, it functions as the substrate recognition component of an E3-ubiquitin ligase complex that targets hypoxia inducible factor 1α (HIF-1α) for ubiquitination and destruction (16). pVHL regulates the stability of microtubules (17), primary cilium maintenance (18), cell differentiation (19), NF-κB activity (20), and Wnt signaling (21). Besides HIF-1α, pVHL has been found to bind to a dozen different proteins, such as atypical protein kinase C (22), hydroxylated α-chain of collagen IV (23), fibronectin (24), and Pax2 (25). Here, we report that pVHL associates with pre-ribosomes by binding the 40S ribosomal protein S3 (RPS3) and suppresses ribosome biogenesis, leading to inhibition of protein synthesis. Our results unravel a new property of pVHL and implicate it in the regulation of protein synthesis. As well, these findings provide novel insight into the regulation of ribosome biogenesis by a tumor suppressor gene product.
MATERIALS AND METHODS
Cell Culture
Human renal carcinoma 786-O and A498 cells were maintained in RPMI 1640. Human cervical cancer HeLa cells and human embryonic kidney 293T cells were cultured in DMEM. All media were supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. The cells were cultured at 37 °C in 5% CO2 incubator.
Construction of Expression Vectors
The genes encoding the ribosomal proteins were obtained by reverse transcription-polymerase chain reaction and cloned into pcDNA3.1-myc vector. The truncated pVHL was cloned with a C-terminal Myc tag into pcDNA3.1. The vectors encoding HA-VHL-NES and HA-VHL-NESM were generated as described (26, 27). Briefly, HA-VHL-NES and HA-VHL-NESM fusions were produced by fusion of HA-VHL at its C terminus to the strong nuclear export signal (NES) of HIV Rev, LPPLERLTL, or to a mutated form of NES (NESM), LPPRERLTL (the underlined amino acid indicates the mutated change). Non-ribosome factors hNob1, hLtv1, and hEnp1 were cloned into pcDNA3.0, pcDNA3.1, or pCMVTag2B. Vectors encoding glutathione S-transferase (GST) tag or His tag fusion proteins were constructed by inserting PCR-generated fragments into pGEX4T1 or pET28a. The Escherichia coli BL21-Gold(DE3)pLysS cells were transformed with pGEX4T1 or pET28a expression vectors and treated with 0.1 mm isopropyl d-thiogalactoside for 4 h at 37 °C. Bicistronic luciferase reporter plasmids pGL3-Ren-CrPV-FF(Ren/CrPV-FF) and pcDNA-Ren-HCV-FF(Ren/HCV-FF) were as described (28).
Transfection
Transient transfection was performed using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. To obtain stable transfectants, the cells were transfected and 24 h later were switched to medium containing G418 (600 μg/ml), which was changed every 2–3 days. After 3 weeks, G418-resistant stable monoclonal cell lines were chosen for further study.
Western Blot and Immunoprecipitation
Extracts were prepared by lysing cells on ice for 30 min in RIPA buffer (100 mm Tris, 150 mm NaCl, 1% Triton, 1% deoxycholic acid, 0.1% SDS, 1 mm EDTA, and 2 mm NaF, pH 7.5) supplemented with 1 mm sodium vanadate, 1 mm leupeptin, 1 mm aprotinin, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and 1 mm pepstatin A. The supernatant was collected after centrifugation at 12,000 × g for 15 min. Equal amounts of protein were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Proteins of interest were detected by Western blot using specific antibodies. Immunoprecipitation was performed as follows: 500 μg of cell lysates were incubated with 1 μg of primary antibody at 4 °C for 3 h. Twenty microliters of protein A/G-agarose beads was added and the incubation continued overnight. The beads were washed with ice-cold cell lysis buffer and boiled in SDS-PAGE loading buffer for 5 min before electrophoresis. Antibodies against pVHL, RPS3, RPL11, and RPS6 were from Cell Signaling. HA, nNob1, hEnp1/bystin-like, and Myc antibodies were purchased from Santa Cruz Biotechnology. β-Actin and FLAG antibodies were from Sigma.
GST Pulldown Assay
The bacterial cells were lysed using the following buffer: 20 mm Tris-Cl, 2 mm EDTA, 150 mm NaCl, and 0.5% Nonidet P-40, pH 7.5. To monitor the interaction between pVHL and ribosomal proteins, bacterial lysates containing GST fusion proteins were incubated with glutathione-Sepharose 4B beads at 4 °C for 1 h. The beads were washed and incubated with His-tagged proteins. After washing, pVHL and the bound ribosomal proteins were eluted from the beads and subjected to electrophoresis.
Ribosome Profile
For sucrose density gradient fractionation of ribosomes, cells were scraped and collected after adding cycloheximide (100 μg/ml) in culture for 5 min. Cells were then lysed using 0.5% Nonidet P-40 lysis buffer containing 100 mm KCl, 10 mm MgCl2, 100 μg/ml of cycloheximide, 100 units/ml of RNasin, 1 mm DTT and protease inhibitors (1 mm leupeptin, 1 mm pepstatin A, and 1 mm phenylmethylsulfonyl fluoride) for 15–20 min at 4 °C. The lysates were centrifuged at 8,000 × g for 5 min and the supernatants were collected. Equivalent A254 units of total lysate was loaded on a 10–45% linear sucrose gradient and centrifuged at 36,000 × g for 3 h at 4 °C using a Beckman SW40Ti rotor. Fractions were collected and absorbance at A254 was monitored.
Monitoring Protein Synthesis
For monitoring cap-dependent and cap-independent translations, cells were transiently transfected with the bicistronic luciferase reporter and pVHL expression plasmids. After harvesting, luciferase light units were measured using a dual luciferase assay kit following the manufacturer's recommendation (Promega). For monitoring global protein synthesis, an equal number of 786-O and A498 cells were incubated in methionine-free DMEM for 45 min, followed by addition of 10 μCi/well of [35S]methionine (PerkinElmer Life Sciences) for 60 min. Whole cell lysates were prepared and 30–40 μg of protein were loaded onto a 10% SDS-polyacrylamide gel. The proteins resolved on the gel were transferred to a nitrocellulose membrane and 35S-labeled proteins were visualized by autoradiography.
Short Interference RNA (siRNA)
The VHL siRNA oligonucleotides (siVHL-1 and 2) and control siRNA oligonucleotides were designed as described (25) and purchased from GenePharma (Shanghai, China). The sense sequences of these oligonucleotides were: control, 5′-UUCUCCGAACGUGUCACGUTT-3′; siVHL-1, 5′-GCCAGUGUAUACUCUGAAATT-3′; and siVHL-2, 5′-GCUCUACGAAGAUCUGGAATT-3′.
Northern Blot
Total RNA was isolated using TRIzol reagent and Northern blot was performed using the DIG Northern Starter Kit as per the manufacturer's instructions (Roche Applied Science). In brief, 10 μg of RNA was resolved by formamide-agarose gel electrophoresis and blotted onto a Hybond-N membrane. Pre-hybridization and hybridization with digoxigenin-labeled oligonucleotides was carried out overnight at 60 °C in hybridization buffer. After hybridization, the blot was washed twice with 2× SSC buffer, and twice in 0.1× SSC for 30 min. Both buffers contained 0.1% SDS. The membrane was incubated 30 min with anti-DIG-AP and then exposed to x-ray film. The sequences of the probes used for Northern blot analysis are: 5′-ITS1, 5′-DIG-CCTCGCCCTCCGGGCT CCGTTAATTGATC-3′; 18S, 5′-DIG-AT CGGCCCGAGGTTATCTAGAGTCACCAAA-3′; 28S, 5′-DIG-CCTCTTCGGGGGACGCGCGCGTGGCCCCGA-3′.
Immunofluorescence Staining
Cells were grown on glass coverslips, washed with phosphate-buffered saline (PBS), and fixed using formaldehyde (4%) for 20 min, followed by permeabilization with 0.1% Triton X-100. After washing with PBS, the coverslips were blocked in 3% BSA for 1 h, followed by an incubation with primary antibody (in 3% BSA) at 4 °C overnight. The coverslips were washed and incubated with fluorescence dye-conjugated secondary antibody for 30 min. Finally, the cells were stained with DAPI for 3 min in the dark. The stained cells were mounted and observed using a Zeiss LSM 510 META (Axiovert 200) microscope.
RESULTS
pVHL Binds Ribosomal Protein RPS3
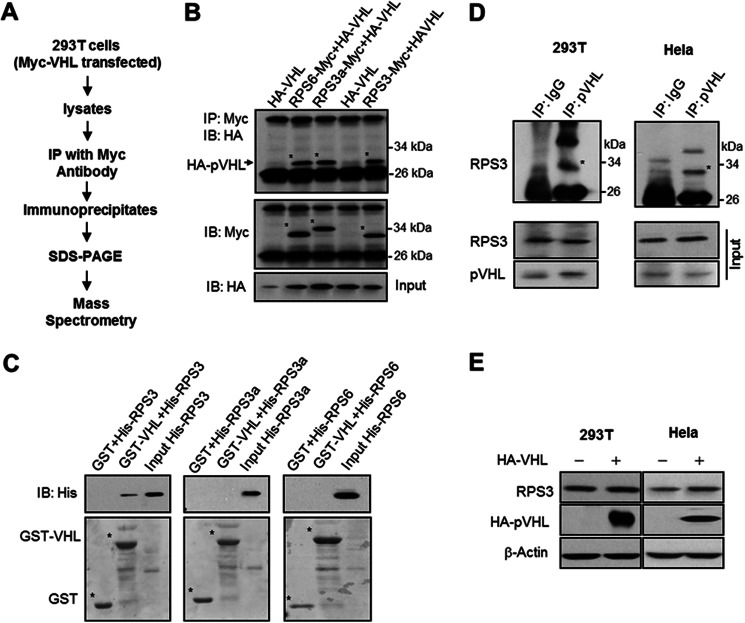
To identify pVHL-interacting proteins, we purified pVHL from 293T cells engineered to produce Myc-tagged pVHL. pVHL was pulled down from cell extracts using an anti-Myc antibody. The immunoprecipitates were resolved on SDS-polyacrylamide gels, and co-purifying proteins were collected and sent for mass spectrometry (MS) analysis (Fig. 1A). Among the peptides identified, we noted the presence of ribosomal proteins, containing both RPSs and 60S subunit ribosomal proteins (RPLs) (supplemental Table S1). Herein, we focus on characterizing the relationship between pVHL and these ribosomal components.
FIGURE 1.
pVHL associates with RPS3. A, schematic diagram illustrating the strategy undertaken to identify pVHL-interacting proteins. B, Western blot analysis validating the interaction between pVHL and several ribosomal proteins. 293T cells were transfected with vectors encoding HA-tagged pVHL and/or Myc-tagged ribosomal proteins as indicated. Following the preparation of cell extracts, Myc-tagged RPS3, RPS3a, and RPS6 were immunoprecipitated (IP) using an anti-Myc antibody and the presence of HA-pVHL was analyzed by immunoblotting (IB) with an anti-HA antibody. C, His-tagged ribosomal proteins were incubated with GST or GST-VHL fusion protein. Following affinity purification, the bound proteins were subjected to SDS-PAGE and Western blot analysis performed using an anti-His antibody. D, interaction between endogenous pVHL and RPS3. Cellular extracts were prepared from HeLa and 293T cells and used in immunoprecipitation experiments with an anti-pVHL antibody. Following fractionation of the immunoprecipitates by SDS-PAGE, immunoblotting was performed using an anti-RPS3 antibody. The top panels represent the anti-pVHL immunoprecipitated samples, whereas the bottom panels denote the corresponding input material. E, overexpression of pVHL does not inhibit RPS3 expression. HeLa and 293T cells were transfected with HA-VHL plasmid. After 24 h, the cells were harvested and protein levels of RPS3 were determined by Western blot.
We first validated that pVHL interacted with ribosomal protein. RPS3 has the highest relative abundance among the identified ribosomal proteins in MS, we therefore detected whether pVHL associated with RPS3. Other proteins such as RPS3a and RPS6 were also examined. 293T cells were co-expressed with HA-tagged pVHL and Myc-tagged versions of RPS3, RPS3a, and RPS6. The results indicate that HA-pVHL associates with Myc-RPS3, Myc-RPS3a, and Myc-RPS6 (Fig. 1B). To determine whether any of these interactions were direct, we performed in vitro pulldown assays with GST or GST-VHL (Fig. 1C). Under our conditions, only His-RPS3 bound GST-pVHL (Fig. 1C). Given that these results suggest a direct interaction between pVHL and RPS3, we probed for, and detected such an association between the endogenous protein pairs in both 293T and HeLa cells (Fig. 1D). Overexpression of pVHL did not reduce the protein level of RPS3 (Fig. 1E), but decreased that of HIF-1α, a well known target of pVHL (supplemental Fig. S1), implying that pVHL does not target RPS3 for destruction. In addition, we found that the β domain of pVHL was sufficient for mediating the pVHL-RPS3 interaction (supplemental Fig. S2).
pVHL Associates with Pre-ribosomes
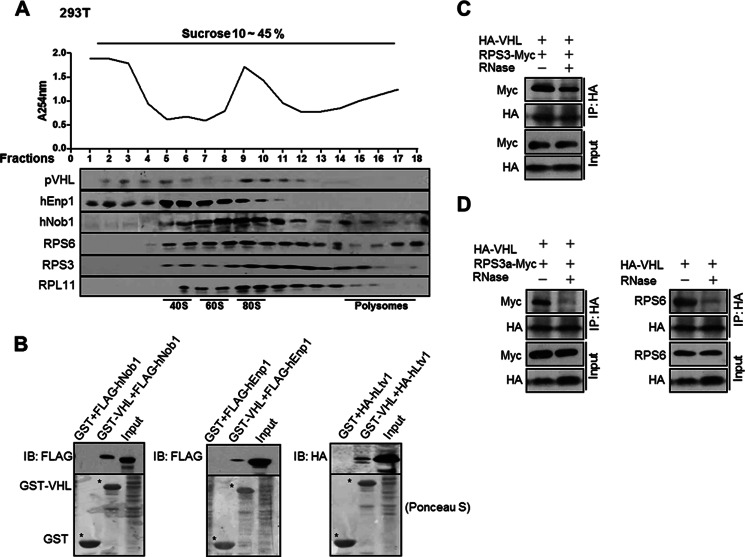
The above results suggested that pVHL interacts with components of the 40S subunits. We then examined whether pVHL was associated with ribosomal complexes. 293T cell extract was centrifuged on a sucrose gradient followed by a Western blot (Fig. 2A). pVHL co-sedimented with 40S complexes (40S and pre-40S subunits) and 80S complexes (80S and pre-90S ribosome). pVHL was not in fractions containing polysomes (fractions 14–18), suggesting that pVHL is not associated with the mature translating ribosomes. Similar results were obtained in HeLa cells (supplemental Fig. S3A). We immunoprecipitated pVHL from pooled fractions 1–4 (non-ribosomal fractions) and fractions 9–12 (80S/90S-conaining fractions) and probed for the presence of RPS3 and RPS6. The results show that pVHL was associated with RPS3 and RPS6 when in 80S/90S-containing fractions (supplemental Fig. S3B). These provide more evidence that pVHL associates with ribosomal complexes.
FIGURE 2.
pVHL associates with pre-ribosomes. A, pVHL is co-sedimented with pre-ribosomes. 293T cell extracts were fractionated by sucrose gradient centrifugation as described under “Materials and Methods.” Each fraction was monitored at A254 (upper panel). The presence of pVHL, hNob1, hEnp1, RPS3, RPS6, and RPL11 in each fraction was determined by Western blot (lower panel). B, pVHL associates with pre-ribosome factors. GST-pVHL proteins were produced in E. coli. The pre-ribosome proteins FLAG-hNob1, HA-hLtv1, or FLAG-hEnp1 were expressed in 293T cells. The bacterial lysates containing GST-pVHL were incubated with glutathione-Sepharose 4B beads at 4 °C for 1 h. The beads were then washed and incubated with 293T cell lysates containing FLAG-hNob1, HA-hLtv1, or FLAG-hEnp1 at 4 °C overnight. The beads were washed and boiled in SDS-PAGE loading buffer for 5 min. The supernatants were collected and resolved on SDS-PAGE. C, ribosome disruption does not abolish the association between pVHL and RPS3. 293T cells were transfected with the indicated expression plasmids. After 24 h, cells were harvested and extracts were treated with or without RNase (5 units, 40 min), followed by immunoprecipitation with an anti-HA antibody. Immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. D, 293T cells were transfected with the indicated expression plasmids. After 24 h, the cells were harvested and cellular proteins were prepared, followed by treatment with RNase as described above.
The hEnp1 (bystin-like) and hNob1 are human homologues of Saccharomyces cerevisiae Enp1p and Nob1 that are components of pre-40S and pre-90S particles in yeast (29–34). Location of hEnp1 and hNob1 was determined and the results show that both proteins and pVHL were sedimented in fractions 5–6 and 9–11 (Fig. 2A). The hEnp1 and hNob1 do not display the same profiles as each other. Nob1 was found in polysome-containing fractions (Fig. 2A). It is reported that the immature small ribosomal subunits can engage in translation initiation and the putative endonuclease Nob1, which is specific to late pre-40S particles can interact with polysomes (35, 36). This is one possible reason why Nob1 was detected in polysome fractions.
Meanwhile, we found that pVHL was associated with hEnp1 and hNob1 (Fig. 2B). The pVHL was also found to interact with another pre-ribosomal protein hLtv1 (human homologue of S. cerevisiae Ltv1p) (29–34) (Fig. 2B). Notably, hNob1, hEnp1, and hLtv1 are implicated in ribosome assembly but not part of mature ribosomes because they are released in the cytoplasm before ribosome maturation is completed (29–34). Altogether, the results suggest that pVHL is present in 40S and 90S precursors.
RNase can degrade ribosomal RNA and disrupt the ribosomal complex. We found that RNase treatment did not affect the interaction between pVHL and RPS3 (Fig. 2C), but abolished the interaction between pVHL and RPS3a or RPS6 (Fig. 2D). These results indicate that the association between pVHL and ribosomal precursors is through the stable interaction with RPS3.
pVHL Inhibits Ribosome Biogenesis
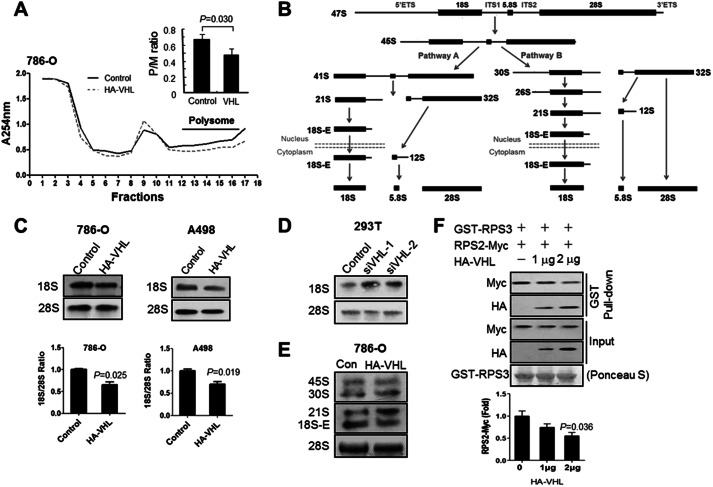
We were interested in assessing the function of pVHL binding to ribosomes. Given the association of pVHL with pre-ribosomal complexes, we asked whether pVHL could be playing a role in ribosome biogenesis. To address this, we took advantage of VHL-null renal carcinoma 786-O cells. The cells were transfected with control empty vector or an HA-VHL expression vector, and stable transfectants were selected. Extracts from 786-O(control) and 786-O(VHL) cells were prepared and ribosomal content was assessed by fractionation using sucrose gradient centrifugation. Re-introduction of VHL into 786-O cells led to a reduction in the polysomes (Fig. 3A). We found that the polysome to monosome ratio (P/M) in 786-O(VHL) cells was decreased compared with that in 786-O cells (Fig. 3A). These data suggest that pVHL reduces the amount of mature ribosomes.
FIGURE 3.
pVHL influences ribosome assembly. A, overexpression of pVHL reduces polysomes in 786-O cells. Ribosomes were isolated from 786-O(control) and 786-O(VHL) cells. The isolated ribosomes were fractionated in a sucrose gradient (10–45%) centrifugation. Each fraction was monitored at A254 nm. The polysome to monosome ratio (P/M) was determined as described (47, 48). The data are mean ± S.E. (n = 3). B, the schematic diagram of rRNA processing. C, overexpression of pVHL reduces the amount of 18S but not 28S rRNA. 18S and 28S rRNAs were determined by Northern blot as described under “Materials and Methods.” Data are mean ± S.E. (n = 3). D, knockdown of pVHL enhanced 18S rRNA level. 293T cells were transfected with the indicated siRNAs and harvested after 48 h. 18S and 28S rRNAs were determined by Northern blot. E, overexpression of pVHL increases the amount of 21S pre-rRNA. 45S, 30S, 21S, and 18S-E rRNA were determined as described above using the 5′-ITS1 probe. F, pVHL inhibits the interaction between RPS3 and RPS2 in a dose-dependent manner. 293T cells were transfected with Myc-RPS2 and different amounts of HA-VHL plasmid. After 24 h, cells were harvested and lysates were prepared. Bacterial lysates containing GST-RPS3 fusion protein were incubated with glutathione-Sepharose 4B beads at 4 °C for 1 h. The beads were washed and incubated with 293T cell extracts at 4 °C overnight. The beads were washed, boiled in SDS-PAGE loading buffer, and resolved on SDS-PAGE. Immunoblotting (IB) was performed with the indicated antibodies. A bar graph shows the fold-difference between the samples. The data are mean ± S.E. (n = 3).
To know whether pVHL affects ribosome maturation, we performed Northern blot analysis to determine the quantity of rRNAs. In mammals, the 40S subunit is composed of 18S rRNA, whereas the 60S subunit is made up of the 28S, 5.8S, and 5S rRNAs. These rRNAs are first synthesized as precursor rRNA (pre-rRNA) molecules that undergo extensive processing to produce mature and functional rRNA (Fig. 3B) (37, 38). We examined pre-rRNA processing by using probes directed against the 18S rRNA, 28S rRNA, and the 5′-most nucleotides of the ITS1 (5′-ITS1). RNA was isolated from 786-O cells ectopically expressing pVHL and analyzed by Northern blot. The results indicate that levels of 18S rRNA, but not 28S rRNA, in 786-O(VHL) cells are lower than those in 786-O(control) cells (Fig. 3C). A498 (another VHL-null renal carcinoma) cells were examined and similar results were obtained (Fig. 3C). Knockdown of pVHL enhanced 18S rRNA levels in 293T cells (Fig. 3D). Previously, it has been reported that acidosis-dependent nucleolar condensation and restriction of rDNA transcription requires nucleolar pVHL (39). We determined the effect of pVHL on rDNA transcription in our system and the results indicate that overexpression of pVHL had little effect on the level of pre-rRNA (supplemental Fig. S4A). In addition, we found that the 21S precursor rRNAs were accumulated and 18S-E was reduced in 786-O(VHL) cells (Fig. 3E).
Initiation of 18S rRNA processing is strictly dependent on the assembly of a subset of initiation-RPS proteins, such as RPS3a, RPS4, and RPS6. Progression of the nuclear and cytoplasmic maturation steps is dependent on a different set of RPS proteins, including RPS2, RPS15, and RPS21. RPS3 is one of the intermediate associating proteins, which links the early associating initiation-RPS proteins and late-associating progression-RPS proteins (40). Depletion of RPS2 delays pre-rRNA processing and leads to the accumulation of 20S pre-rRNA in yeast (41, 42) and 21S pre-rRNA in human (40). The data of Fig. 3E are consistent with this. In the 40S ribosomal subunits, RPS2 is directly associated with RPS3 (43, 44). We therefore supposed that pVHL might prevent RPS2 from binding to the pre-ribosomal complex. We do find that pVHL suppressed the interaction between RPS3 and RPS2 (Fig. 3F), but not the interaction between RPS3 and the initiation protein RPS3a (supplemental Fig. S4B). We examined whether depletion of RPS2 could lead to reduction of 18S rRNA and found that knockdown of RPS2 reduced 18S rRNA (supplemental Fig. S4C). In addition, we determined the expression of RPS2 by the ribosome profile assay in 786-O(VHL) and control cells. A reduction of RPS2 was observed in the 40S sized fractions of 786-O(VHL) cells (supplemental Fig. S4D). These results suggest that pVHL impairs 40S ribosome biogenesis through suppressing the RPS2-RPS3 interaction.
pVHL Causes Retention of Pre-40S Ribosomal Subunits in the Nucleus
The assembly of the ribosomal subunits is completed in the cytoplasm. Blockade of the ribosome assembly could be arising as a consequence of accumulation of ribosome proteins in the nucleus. As seen in Fig. 3B, most of the 18S precursors are found exclusively in the nucleus, including the 21S pre-rRNA. Moreover, the last precursor 18S-E pre-rRNA is exported from the nucleus and processed to 18S rRNA in cytoplasm (38). To determine whether reduction of 18S-E rRNA in pVHL overexpression cells could lead to the pre-40S subunits retention in the nucleus, we detected subcellular localization of the 40S ribosomal protein RPS3 and pre-40S factor hLtv1. To this end, we utilized expression vectors encoding pVHL, pVHL-NES, and pVHL-NESM. pVHL and pVHL-NESM can freely enter into the nucleus, but pVHL-NES is nuclear-import defective (26, 27). Overexpression of pVHL and pVHL-NESM, but not pVHL-NES, enhanced RPS3 (Fig. 4A) and hLtv1 (Fig. 4B) levels in the nucleus. These results suggest that pVHL leads to nuclear retention of pre-40S precursors. Western blot results confirmed the nuclear accumulation of RPS3 and RPS6 in 786-O cells that stably expressed pVHL (Fig. 4C). Expression of pVHL had little effect on nuclear levels of the 60S subunit ribosomal protein RPL11 (Fig. 4C and supplemental Fig. S5A). Interestingly, the protein levels of RPS3 and RPS6 were found decreased in the cytoplasm (Fig. 4C). However, the total protein levels of RPS3 and RPS6 were not affected (Fig. 4D). The reduction of polysomes described in Fig. 3A could be the consequence of the retention of pre-40S in the nucleus and the lack of 40S subunits in the cytoplasm to form mature 80S ribosomes.
FIGURE 4.
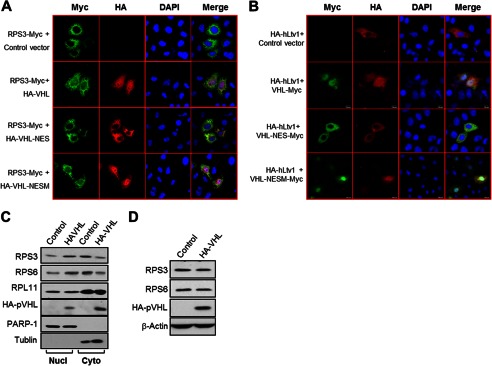
pVHL induces nuclear retention of pre-40S ribosomal subunits. A and B, nuclear expression pVHL promotes RPS3 and hLtv1 nucleus retention. HeLa cells were transfected as indicated. Twenty-four hours post-transfection, the cells were immunostained and visualized under a confocal microscopy. C, nuclear expression of pVHL results in retention of RPS3 and RPS6 in the nucleus. 786-O(VHL) and 786-O(control) stable cells were harvested, lysed, and fractionated into nuclear and cytoplasmic extracts, and analyzed by immunoblotting. D, overexpression of pVHL does not affect the total protein level of RPS3 and RPS6. 786-O stable cells were harvested and the protein levels of RPS3 and RPS6 were determined by Western blot.
We have shown that pVHL suppressed the interaction of RPS3 and RPS2 (Fig. 3F) and RPS2 depletion led to 18S rRNA reduction (supplemental Fig. S4C). We therefore examined whether depletion of RPS2 could also result in pre-40S subunit sequestration. We found that inhibition of RPS2 led to nuclear sequestration of RPS3, RPS6, and hENP1 (supplemental Fig. S5B). Deletion of RPS2 had little effect on RPL11 nuclear and cytoplasm distribution.
pVHL Inhibits Both Cap-dependent and Cap-independent Translation
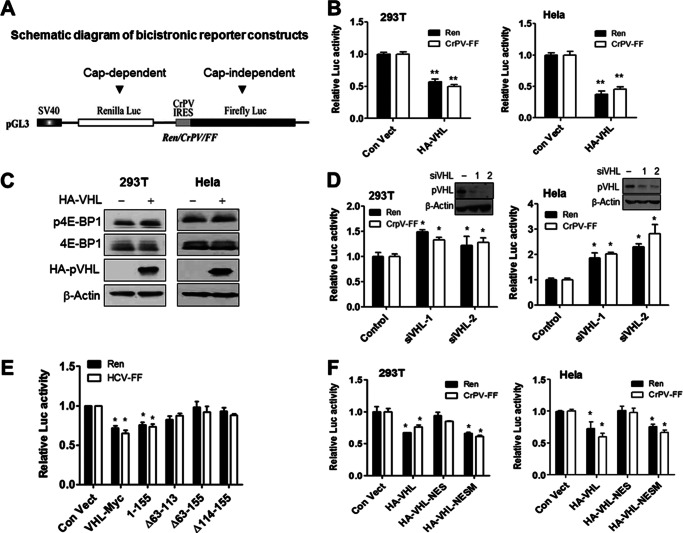
Our results suggest that pVHL suppresses ribosome biogenesis. We next determined the effects of pVHL on global protein synthesis. We therefore assessed the effects of pVHL on translation using the bicistronic luciferase reporter plasmid (Fig. 5A). Overexpression of pVHL inhibited both cap-dependent Renilla luciferase (REN) translation and CrPV internal ribosome entry site-mediated (CrPV IRES-mediated) translation of firefly luciferase (FF) (Fig. 5B). Another luciferase reporter plasmid Ren/HCV/FF (HCV-IRES-mediated) (28) was employed and similar results were obtained (supplemental Fig. S6A). The result was not a consequence of effects on integrity of reporter mRNA (supplemental Fig. S6B). The translation repressor protein 4E-BP1 controls translation initiation (45). We found that pVHL had little effect on expression and phosphorylation of 4E-BP1 (Fig. 5C).
FIGURE 5.
pVHL inhibits both cap-dependent and cap-independent translations. A, schematic diagram of bicistronic reporter constructs used in this study (28). B, ectopic expression of pVHL inhibits cap-dependent and cap-independent translations in vivo. HeLa and 293T cells were transfected with the indicated bicistronic reporter construct (Ren/CrPV/FF) and a pVHL expression plasmid. After 48 h, cells were harvested and lysates were prepared for Renilla and firefly luciferase activity measurements. The data are mean ± S.E. (n = 3). **, p < 0.01 versus control. C, overexpression of pVHL had no effect on 4E-BP1 and phosphor-4E-BP1 levels. 293T and HeLa cells were transfected with HA-VHL and harvested after 24 h. The expression of 4E-BP1 and phosphor-4E-BP1 is analyzed by Western blot. D, suppression of pVHL increases cap-dependent and cap-independent translation. HeLa and 293T cells were transfected with the indicated bicistronic reporter construct (Ren/CrPV/FF) and VHL or control (non-targeting) siRNAs. After 48 h, the cells were harvested for luciferase activity measurement. Data represent mean ± S.E. (n = 3). *, p < 0.05 versus control. The inset Western blot indicates levels of pVHL suppression achieved by RNA interference. E, the β domain of pVHL is required for inhibition of translation. 293T cells were transfected with the bicistronic reporter plasmid (HCV/FF) and the indicated pVHL mutants. After 48 h, cells were harvested for luciferase activity measurements. The data represent mean ± S.E. (n = 3). *, p < 0.05 versus control. F, the subcellular localization of pVHL determines its ability to inhibit translation. 293T and HeLa cells were transfected with the bicistronic reporter construct (Ren/CrPV/FF) and the indicated pVHL expression plasmids. After 48 h, cells were harvested for luciferase activity measurement. The data represent the mean ± S.E. (n = 3). *, p < 0.05 versus control. Ren, Renilla; FF, firefly.
Moreover, suppression of VHL by RNA interference enhanced both cap-dependent and cap-independent protein translations (Fig. 5D and supplemental Fig. S7A). Because RPS3 binds pVHL at the β domain (supplemental Fig. S2), we assessed if this domain was also responsible for the observed inhibition of translation. We found that pVHL and pVHL(1–155) inhibited translation (Fig. 5E). However, pVHL(Δ63–113), pVHL(Δ63–155), and pVHL(Δ114–155) had little effect on translation (Fig. 5E). These mutations had little effect on the nucleus location of pVHL (supplemental Fig. S8). We also assessed whether altered subcellular localization of pVHL would have an effect on its ability to inhibit protein synthesis. The results show that pVHL and pVHL-NESM, but not pVHL-NES, inhibit translation (Fig. 5F and supplemental Fig. S7B).
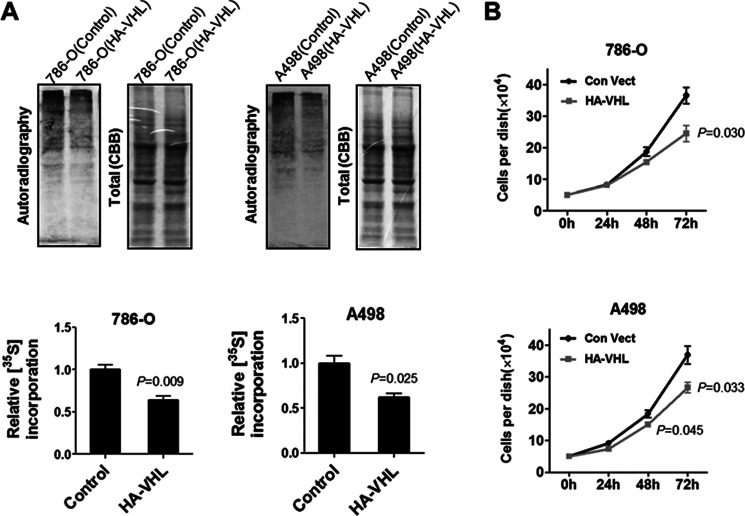
Next, we determined whether pVHL was exerting an inhibition effect on global protein synthesis by performing [35S]methionine labeling of cells lacking or expressing wild-type pVHL (Fig. 6A). We noted that re-introduction of VHL into 786-O and A498 cells repressed global protein synthesis (Fig. 6A). Consistent with this, re-expression of VHL in 786-O and A498 cells attenuated proliferation of both cells (Fig. 6B).
FIGURE 6.
pVHL inhibits protein synthesis. A, pVHL expression inhibits protein synthesis. 786-O and A498 cells that were stably transfected with HA-VHL were seeded in 12-well plates. The next day, the medium was removed and methionine-free medium was added. After 45 min, the cells were provided with [35S]methionine (10 μCi/ml) and incubated for 60 min. The cells were harvested and lysed. Cellular proteins were resolved on SDS-PAGE and the 35S-labeled proteins were visualized by autoradiography. 786-O and A498 cells stably transfected with control vector were used as control. The 35S-incorporation was determined by measuring the density of the 35S-labeled protein (autoradiograph) and normalized to that of total proteins (Coomassie Blue stain). The data are mean ± S.E. (n = 3). B, re-expression of pVHL inhibits cell proliferation. 786-O and A498 cells stably expressing pVHL were selected as described under “Materials and Methods.” Cell proliferation was determined by counting the cell numbers using a hemocytometer. The data are mean ± S.E. (n = 3).
DISCUSSION
Ribosome production is enhanced in cancer cells and the enhanced ribosome production may facilitate tumorigenesis (4, 5, 9). The mechanisms of up-regulation of ribosome biogenesis in cancer cells remain unclear. Several tumor suppressors and oncogenes have been found to influence the formation of ribosomes. pRB and p53 influence ribosome biogenesis through inhibiting rRNA synthesis and RNA polymerases transcription (6–9). PTEN represses transcription of polymerase I and MYC increases transcription of polymerase I, II, and III (10). In this article, we demonstrate that pVHL inhibits ribosome biogenesis through interfering with the assembly of ribosomes.
As an adaptor protein, pVHL interacts with many proteins (15). The identification of interacting proteins has helped to elucidate the multiple functions of pVHL. For example, the function of pVHL to block assembly of the extracellular fibronectin matrix has been linked to its ability to bind fibronectin (24). We find that RPS3 is a new interacting protein of pVHL. In this report we provide data that suggests that pVHL binds RPS3 and interferes with ribosome assembly. Besides RPS3, other ribosomal proteins were also identified in our MS assay (supplemental Table S1). In ribosome biogenesis, the 90S precursor contains the processed versions of ribosomal proteins (RPSs and RPLs) and RPS3 was identified in 90S precursors (31). pVHL directly binds RPS3 and therefore it can associate with the 90S precursors. So, it is expected that pVHL may have indirect interaction with other proteins in 90S particles.
pVHL is co-sedimented with pre-40S ribosomal subunits and impaired 18S rRNA processing (Figs. 2 and 3C), indicating that pVHL inhibits biogenesis of ribosomal small subunits. RPS2 is a progression-RPS protein that is necessary for nuclear export and cytoplasmic maturation steps (38). We found that depletion of RPS2 reduced 18S rRNA (supplemental Fig. S4D) and led to nuclear sequestration of RPS3, RPS6, and hEnp1, which indicates the nuclear retention of pre-40S subunits (supplemental Fig. S5A). Overexpression of pVHL prevents the interaction between RPS3 and RPS2 (Fig. 3F), leading to 18S rRNA reduction (Fig. 3) and nuclear retention of pre-40S subunits (Fig. 4). These results suggest that pVHL suppresses 40S subunit assembly by preventing the RPS3-RPS2 interaction. pVHL had little effect on the 28S rRNA level (Fig. 3, C and D) and RPL11 cellular location (Fig. 4C and supplemental Fig. S5). Thus, pVHL might not influence 60S ribosomal subunits.
Overexpression of pVHL reduced polysomes (Fig. 3, supplemental Fig. S4D). Consistent with these results, pVHL was found to inhibit translation (Fig. 5, B and D, and supplemental Figs. S6A and S7A). Finally, we demonstrate that expression of pVHL inhibits protein synthesis in 786-O and A498 cells (Fig. 6). Taken together, our results suggest that pVHL inhibits global translation.
Ribosome biogenesis is an important process for a proliferating cell (46). The biological mechanisms that regulate cell proliferation also control ribosome biogenesis and have been linked to transformation. Up-regulation of ribosome biogenesis is associated with an increased risk of tumor onset and elevated ribosome biogenesis is associated with acquisition of the neoplastic phenotype (9). As a result of oncogene gain-of-function mutations or tumor suppressor loss, ribosome biogenesis is frequently enhanced and associated with a cellular growth advantage, although the mechanisms are unclear. We report here that pVHL inhibits ribosome biogenesis through ribosomal protein assembly. pVHL is a tumor suppressor and its loss is associated with tumor initiation and progression (13). Our results may uncover a new function of pVHL in regulation of tumor cell proliferation by controlling the ribosome biogenesis.
This work was supported by Natural Science Foundation of China Grants 30970586 and 31270829 and the Innovation Program of Chinese Academy of Sciences Grant KSCX2-EW-R-09.

This article contains supplemental Table S1 and Figs. S1–S8.
- VHL
- von Hippel-Landau
- HIF-1α
- hypoxia inducible factor 1α
- NES
- nuclear export signal
- RPS
- 40S ribosomal protein
- DIG
- digoxigenin.
REFERENCES
- 1. Lafontaine D. L., Tollervey D. (2001) The function and synthesis of ribosomes. Nat. Rev. Mol. Cell. Biol. 2, 514–520 [DOI] [PubMed] [Google Scholar]
- 2. Fatica A., Tollervey D. (2002) Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318 [DOI] [PubMed] [Google Scholar]
- 3. Albanèse V., Reissmann S., Frydman J. (2010) A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 189, 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belin S., Beghin A., Solano-Gonzàlez E., Bezin L., Brunet-Manquat S., Textoris J., Prats A. C., Mertani H. C., Dumontet C., Diaz J. J. (2009) Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One 4, e7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruggero D., Pandolfi P. P. (2003) Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 6. Treré D., Ceccarelli C., Montanaro L., Tosti E., Derenzini M. (2004) Nucleolar size and activity are related to pRb and p53 status in human breast cancer. J. Histochem. Cytochem. 52, 1601–1607 [DOI] [PubMed] [Google Scholar]
- 7. Cavanaugh A. H., Hempel W. M., Taylor L. J., Rogalsky V., Todorov G., Rothblum L. I. (1995) Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374, 177–180 [DOI] [PubMed] [Google Scholar]
- 8. Brandenburger Y., Jenkins A., Autelitano D. J., Hannan R. D. (2001) Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J. 15, 2051–2053 [DOI] [PubMed] [Google Scholar]
- 9. Montanaro L., Treré D., Derenzini M. (2012) Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim. Biophys. Acta 1825, 101–110 [DOI] [PubMed] [Google Scholar]
- 10. van Riggelen J., Yetil A., Felsher D. W. (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309 [DOI] [PubMed] [Google Scholar]
- 11. Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003) Ribosome assembly in eukaryotes. Gene 313, 17–42 [DOI] [PubMed] [Google Scholar]
- 12. Milkereit P., Kühn H., Gas N., Tschochner H. (2003) The pre-ribosomal network. Nucleic Acids Res. 31, 799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaelin W. G. (2007) Von Hippel-Lindau disease. Annu. Rev. Pathol. 2, 145–173 [DOI] [PubMed] [Google Scholar]
- 14. Kim W. Y., Kaelin W. G. (2004) Role of VHL gene mutation in human cancer. J. Clin. Oncol. 22, 4991–5004 [DOI] [PubMed] [Google Scholar]
- 15. Frew I. J., Krek W. (2008) pVHL. A multipurpose adaptor protein. Sci. Signal. 1, pe30. [DOI] [PubMed] [Google Scholar]
- 16. Kaelin W. G. (2008) The von Hippel–Lindau tumour suppressor protein. O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 [DOI] [PubMed] [Google Scholar]
- 17. Hergovich A., Lisztwan J., Barry R., Ballschmieter P., Krek W. (2003) Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 5, 64–70 [DOI] [PubMed] [Google Scholar]
- 18. Thoma C. R., Frew I. J., Hoerner C. R., Montani M., Moch H., Krek W. (2007) pVHL and GSK3β are components of a primary cilium maintenance signalling network. Nat. Cell Biol. 9, 588–595 [DOI] [PubMed] [Google Scholar]
- 19. Davidowitz E. J., Schoenfeld A. R., Burk R. D. (2001) VHL induces renal cell differentiation and growth arrest through integration of cell-cell and cell-extracellular matrix signaling. Mol. Cell. Biol. 21, 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang H., Minamishima Y. A., Yan Q., Schlisio S., Ebert B. L., Zhang X., Zhang L., Kim W. Y., Olumi A. F., Kaelin W. G. (2007) pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-κB agonist Card9 by CK2. Mol. Cell 28, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chitalia V. C., Foy R. L., Bachschmid M. M., Zeng L., Panchenko M. V., Zhou M. I., Bharti A., Seldin D. C., Lecker S. H., Dominguez I., Cohen H. T. (2008) Jade-1 inhibits Wnt signaling by ubiquitilating β-catenin and mediates Wnt pathway inhibition by pVHL. Nat. Cell Biol. 10, 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okuda H., Saitoh K., Hirai S., Iwai K., Takaki Y., Baba M., Minato N., Ohno S., Shuin T. (2001) The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J. Biol. Chem. 276, 43611–43617 [DOI] [PubMed] [Google Scholar]
- 23. Grosfeld A., Stolze I. P., Cockman M. E., Pugh C. W., Edelmann M., Kessler B., Bullock A. N., Ratcliffe P. J., Masson N. (2007) Interaction of hydroxylated collagen IV with the von Hippel-Lindau tumor suppressor. J. Biol. Chem. 282, 13264–13269 [DOI] [PubMed] [Google Scholar]
- 24. Ohh M., Yauch R. L., Lonergan K. M., Whaley J. M., Stemmer-Rachamimov A. O., Louis D. N., Gavin B. J., Kley N., Kaelin W. G., Jr., Iliopoulos O. (1998) The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol. Cell 1, 959–968 [DOI] [PubMed] [Google Scholar]
- 25. Yan B., Jiao S., Zhang H. S., Lv D. D., Xue J., Fan L., Wu G. H., Fang J. (2011) Prolyl hydroxylase domain protein 3 targets Pax2 for destruction. Biochem. Biophys. Res. Commun. 409, 315–320 [DOI] [PubMed] [Google Scholar]
- 26. Lee S., Neumann M., Stearman R., Stauber R., Pause A., Pavlakis G. N., Klausner R. D. (1999) Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 19, 1486–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis M. D., Roberts B. J. (2003) Role of nuclear and cytoplasmic localization in the tumour-suppressor activity of the von Hippel-Lindau protein. Oncogene 22, 3992–3997 [DOI] [PubMed] [Google Scholar]
- 28. Bordeleau M. E., Robert F., Gerard B., Lindqvist L., Chen S. M., Wendel H. G., Brem B., Greger H., Lowe S. W., Porco J. A., Jr., Pelletier J. (2008) Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 118, 2651–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Campbell M. G., Karbstein K. (2011) Protein-protein interactions within late pre-40S ribosomes. PLoS One 6, e16194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carron C., O'Donohue M. F., Choesmel V., Faubladier M., Gleizes P. E. (2011) Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 39, 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., Gavin A. C., Hurt E. (2002) 90S pre-ribosomes include the 35S-pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10, 105–115 [DOI] [PubMed] [Google Scholar]
- 32. Granneman S., Petfalski E., Swiatkowska A., Tollervey D. (2010) Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 29, 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. (2003) The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zemp I., Wild T., O'Donohue M. F., Wandrey F., Widmann B., Gleizes P. E., Kutay U. (2009) Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185, 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soudet J., Gélugne J. P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A. (2010) Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 29, 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strunk B. S., Loucks C. R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C. L., 3rd, Karbstein K., Skiniotis G. (2011) Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333, 1449–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hadjiolova K. V., Nicoloso M., Mazan S., Hadjiolov A. A., Bachellerie J. P. (1993) Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 212, 211–215 [DOI] [PubMed] [Google Scholar]
- 38. Rouquette J., Choesmel V., Gleizes P. E. (2005) Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 24, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mekhail K., Rivero-Lopez L., Khacho M., Lee S. (2006) Restriction of rRNA synthesis by VHL maintains energy equilibrium under hypoxia. Cell Cycle 5, 2401–2413 [DOI] [PubMed] [Google Scholar]
- 40. O'Donohue M. F., Choesmel V., Faubladier M., Fichant G., Gleizes P. E. (2010) Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190, 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perreault A., Bellemer C., Bachand F. (2008) Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 36, 6132–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferreira-Cerca S., Pöll G., Gleizes P. E., Tschochner H., Milkereit P. (2005) Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 20, 263–275 [DOI] [PubMed] [Google Scholar]
- 43. Ferreira-Cerca S., Pöll G., Kühn H., Neueder A., Jakob S., Tschochner H., Milkereit P. (2007) Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell 28, 446–457 [DOI] [PubMed] [Google Scholar]
- 44. Bulygin K., Chavatte L., Frolova L., Karpova G., Favre A. (2005) The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30, and S15. Biochemistry 44, 2153–2162 [DOI] [PubMed] [Google Scholar]
- 45. Hay N., Sonenberg N. (2004) Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 46. Schmidt E. V. (1999) The role of c-myc in cellular growth control. Oncogene 18, 2988–2996 [DOI] [PubMed] [Google Scholar]
- 47. Bordeleau M. E., Mori A., Oberer M., Lindqvist L., Chard L. S., Higa T., Belsham G. J., Wagner G., Tanaka J., Pelletier J. (2006) Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2, 213–220 [DOI] [PubMed] [Google Scholar]
- 48. Nemoto N., Singh C. R., Udagawa T., Wang S., Thorson E., Winter Z., Ohira T., Ii M., Valásek L., Brown S. J., Asano K. (2010) Yeast 18S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J. Biol. Chem. 285, 32200–32212 [DOI] [PMC free article] [PubMed] [Google Scholar]