TABLE 1.
Data collection and refinement statistics for BACOVA_00430 (Protein Data Bank code 3ohg)
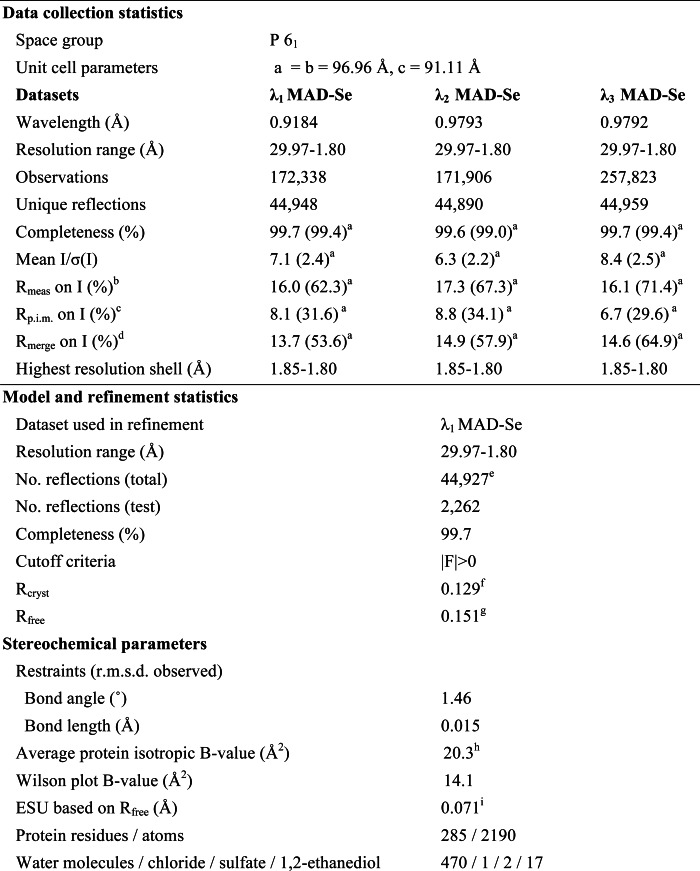
a Highest resolution shell.
b Rmeas (redundancy-independent Rmerge) = Σhkl(n/(n − 1))1/2 Σ|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl) (24), where n is the number of observations of a given reflection.
c Rp.i.m. (precision-indicating Rmerge) = Σhkl((1/(n − 1))1/2 Σi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl) (25, 26).
d Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl).
e Typically, the number of unique reflections used in refinement is slightly less than the total number that were integrated and scaled. Reflections are excluded due to negative intensities and rounding errors in the resolution limits and cell parameters.
f Rcryst = Σ‖Fobs| − |Fcalc‖/Σ |Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
g Rfree as for Rcryst, but for 5.0% of the total reflections chosen at random and omitted from refinement.
h This value represents the total B that includes overall TLS refinement and residual B components.
i ESU, estimated overall coordinate error (23).
