Figure 1.
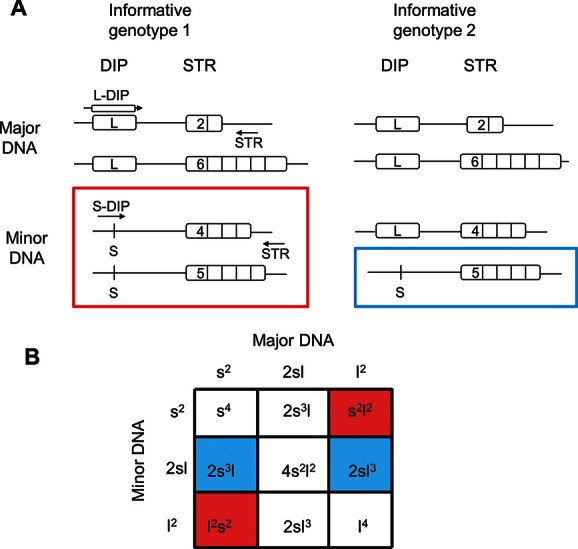
A: DIP–STR informative genotypes. A DIP–STR haplotype is analyzed by using PCR primers overlapping the DIP on one side (either L-DIP or S-DIP primer) and downstream the STR (STR primer) on the other side. When major and minor DNA contributors are opposite homozygous LL/SS or SS/LL (“informative genotype 1”), two minor DNA haplotypes can be identified in the mixture (red box); conversely, when the major DNA contributor is homozygous, either SS or LL, and the minor DNA contributor is heterozygous (“informative genotype 2”), one minor DNA haplotype can be identified in the mixture (blue box). Arrows indicate PCR primers. B: Theoretical evaluation of the occurrence of informative markers. Letter s and l indicate the allele frequencies of S and L alleles, respectively. In red are the probabilities of informative DIP genotypes enabling the identification of two minor DNA haplotypes (“informative genotype 1”). Depending on the linked STR alleles, these are the same or different. In blue are the probabilities of informative DIP genotypes enabling the identification of one minor DNA haplotype (“informative genotype 2”). The sum of these values (s2l2+l2s2+2s3l+2sl3) gives the probability of having any type of informative genotype (I) in a mixture of two DNA.
