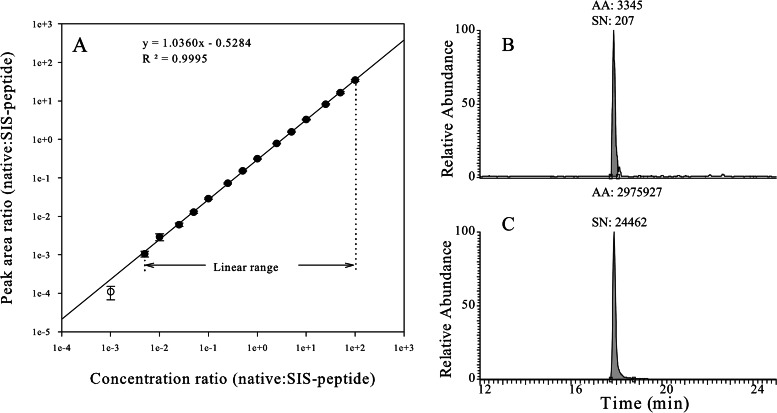
Fig. 1.
Calibration curve for MAVS SID-SRM-MS assay. A stable isotope labeled [12C]-high-responding signature peptide of MAVS with its native flanking sequences (LTK-VSASTVPTDGSSR-NEE) was quantified via amino acid analysis and used as the calibrator to characterize the assay dynamic range. This peptide was trypsin digested in the same matrix as that used for the analysis of A549 cell protein extracts. We diluted the tryptic digest to generate a range of analyte concentrations spanning a 100,000-fold concentration range (from 50 amol to 5 pmol on column). These various analyte concentrations were combined with a constant amount of [13C]-analyte peptide internal standard. Four replicate LC-SRM-MS analyses of each sample dilution were performed in order from most diluted to most concentrated. Linear regression analysis was performed on the observed peak area ratios (native:heavy) versus the concentration ratios (native:heavy) to generate calibration curves. A, the linear regression analysis (1/x weighted) for MAVS illustrates the linear dynamic range of the assay. The error bars indicate the standard deviation of the measurements. B, extract ion chromatogram of the native peptide of MAVS (250 amol on column) in which the x-axis is the chromatographic retention time (min) and the y-axis is the extracted ion intensity. AA, area under the curve; SN, signal-to-noise ratio. C, extract ion chromatogram of the SIS peptide of MAVS.