Abstract
Knowledge representation of the role of phosphorylation is essential for the meaningful understanding of many biological processes. However, such a representation is challenging because proteins can exist in numerous phosphorylated forms with each one having its own characteristic protein–protein interactions (PPIs), functions and subcellular localization. In this article, we evaluate the current state of phosphorylation event curation and then present a bioinformatics framework for the annotation and representation of phosphorylated proteins and construction of phosphorylation networks that addresses some of the gaps in current curation efforts. The integrated approach involves (i) text mining guided by RLIMS-P, a tool that identifies phosphorylation-related information in scientific literature; (ii) data mining from curated PPI databases; (iii) protein form and complex representation using the Protein Ontology (PRO); (iv) functional annotation using the Gene Ontology (GO); and (v) network visualization and analysis with Cytoscape. We use this framework to study the spindle checkpoint, the process that monitors the assembly of the mitotic spindle and blocks cell cycle progression at metaphase until all chromosomes have made bipolar spindle attachments. The phosphorylation networks we construct, centered on the human checkpoint kinase BUB1B (BubR1) and its yeast counterpart MAD3, offer a unique view of the spindle checkpoint that emphasizes biologically relevant phosphorylated forms, phosphorylation-state–specific PPIs and kinase–substrate relationships. Our approach for constructing protein phosphorylation networks can be applied to any biological process that is affected by phosphorylation.
Database URL: http://www.yeastgenome.org/
Introduction
Protein phosphorylation is an essential regulatory mechanism that plays a role in many biological processes. Phosphorylation of a protein may lead to activation or repression of its activity, alternative subcellular localization and interaction with different binding partners. Representation of phosphorylated proteins along with their phosphoform-specific functions and protein–protein interactions (PPIs) is critical for knowledge discovery in many areas of research. Although information about protein phosphorylation and its functional impact is plentiful in the scientific literature and in some curated databases, its integration and representation is lagging behind.
Many aspects of the cell cycle are fundamentally dependent on protein phosphorylation. Our understanding of these processes would benefit greatly from the development of a network representation that takes into account protein phosphorylation and its functional effects. An example of such a process is the spindle checkpoint. In eukaryotes from yeast to humans, the spindle checkpoint ensures the fidelity of chromosome segregation by arresting cells before anaphase until every sister chromatid pair has made bipolar attachments to the mitotic spindle (1).
The spindle checkpoint is complex, involving seven core proteins, namely MAD1L1 (Mad1), MAD2L1 (Mad2), BUB1, BUB1B (BubR1), BUB3, MPS1 and AURKB (Aurora B) in humans, and at least a dozen other proteins (1, 2). The critical target of the spindle checkpoint pathway is CDC20. CDC20 is a component of the anaphase promoting complex/cyclosome (APC/C), a multisubunit ubiquitin ligase that promotes anaphase onset by targeting the anaphase inhibitor, PTTG1 (securin), for degradation by the proteasome. Activated by unattached kinetochores or other spindle defects, the spindle checkpoint response promotes the association of BUB1B, BUB3 and MAD2L1 with CDC20 to form the mitotic checkpoint complex (MCC), which inhibits CDC20 activity. As long as CDC20 activity is inhibited, the APC/C is unable to ubiquitinate PTTG1 and the cell cannot progress from metaphase to anaphase (1, 3). In addition to their roles in cell cycle arrest, some spindle checkpoint components also actively promote the formation of correct microtubule-chromosome attachments (4).
Of the core spindle checkpoint proteins, three—BUB1, MPS1 and AURKB—are protein kinases (3). BUB1B was long assumed to be a protein kinase and several putative substrates have been identified; however, recent work suggests it may in fact be a pseudokinase (5). In addition, all seven checkpoint proteins are phosphorylated, suggesting that protein phosphorylation may play a central role in controlling the checkpoint (3, 6). Indeed, studies have identified numerous phosphorylation events that are critical for a robust spindle checkpoint response (3). The spindle checkpoint is also an extremely active area of research. A recent search in PubMed for just one of the core checkpoint proteins, BUB1, returned 583 documents. Thus, a systematic representation of the vast existing knowledge about the spindle checkpoint and the role that phosphorylation plays in it would be a valuable tool for gaining further insights into the process.
In this article, we assess the current state of phosphorylation event curation, identifying areas that are not adequately covered by existing resources. Next, we use the spindle checkpoint as a case study to demonstrate an integrated approach to the construction of protein phosphorylation networks that addresses these gaps in curation and representation. Our approach combines information retrieved via text mining and data mining of PPI databases and uses PRO (7) as a framework to describe the protein forms and complexes and associate them with attributes such as function and localization. Cytoscape (8) is used to visualize the network and highlight terms with attributes of interest. Specifically, we build a phosphorylation network centered on the human checkpoint protein BUB1B and show that it provides a different, but complementary view of the spindle checkpoint from existing protein interaction network building tools. Finally, we compare the BUB1B network with that of its yeast counterpart, MAD3, and discuss the conservation of the human and yeast spindle checkpoints.
Methods
Analysis of phosphorylation and PPI information in existing resources
Data on substrates, kinases and phosphorylation sites were gathered from the following resources: Phospho.ELM (9), PhosphoSitePlus (10), HPRD (Human Protein Reference Database) (11), PhosphoGRID (12), P3DB (13), PhosPhAt (14,15) and UniProtKB (16). All information that could be associated with a UniProtKB accession number was used in the aggregate analysis.
To determine the number of phosphorylation-related articles in PubMed, we analyzed the >22 million abstracts in PubMed using the text mining tool RLIMS-P (Rule-based LIterature Mining System for Protein Phosphorylation) (17), which identifies articles with phosphorylation information.
The number of human phosphorylation reactions in Reactome (18) was determined using the Reactome advanced search function with the following settings: class = ReactionlikeEvent; input = ATP; output = ADP and species = Homo sapiens.
The number of phosphorylated protein forms in PRO was determined by selecting ‘phosphorylated forms’ from the Quick Links menu in the PRO search tool. To calculate the fraction of multiply phosphorylated forms in PRO, we extracted phosphorylation site information from PRO term definitions and annotation. To find all phosphorylated forms in PRO annotated with PPI information, we searched PRO for terms with ‘phosphorylated’ in the name that were annotated with the GO (19) term GO:0005515 (protein binding) or its child terms. We identified all phosphorylated forms in PRO with functional annotation by searching for phosphorylated form terms containing the relations ‘located_in’ or ‘has_function’ or ‘participates_in’, indicating annotation with GO subcellular location, function and biological process terms, respectively.
The numbers of binding interactions and posttranslational modification (PTM) relationships in the STRING (Search Tool for Retrieval of Interacting Genes/Proteins) (20) were determined from the protein actions file ‘protein.actions.v9.0.txt’ available on the STRING Web site. The BUB1B-centered STRING network was built using the STRING web interface (version 9.05). Interactions were restricted to those that were experimentally observed (as opposed to predicted) by selecting ‘Experiments’ and ‘Databases’ as the active prediction methods.
Overview of the phosphorylation network workflow
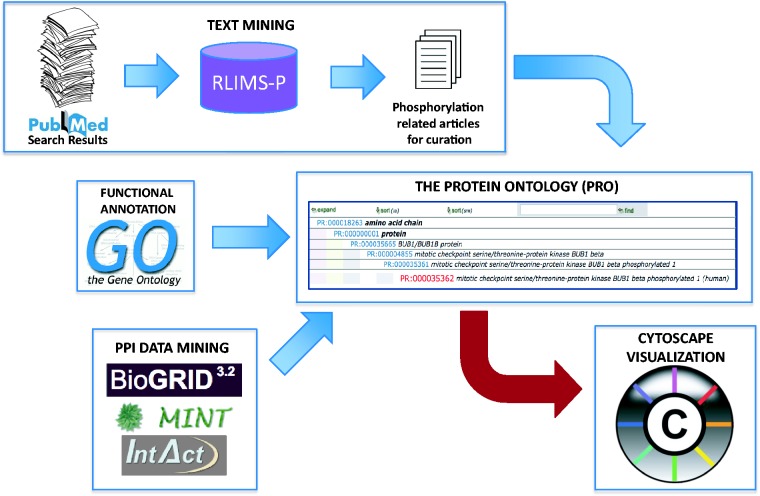
Information about the human spindle checkpoint proteins BUB1 and BUB1B, and their yeast homologs BUB1 and MAD3 was captured through a combination of text and data mining approaches (Figure 1). First, a list of BUB1-, BUB1B- and/or MAD3-related articles was obtained by searching PubMed using the gene name. Then, because of our interest in the role of phosphorylation in the checkpoint, we selected relevant articles for curation using RLIMS-P, a rule-based system specifically designed to extract protein phosphorylation information from text (17, 21). Additional PPIs involving spindle checkpoint proteins were gathered from several curated PPI databases. Information on protein forms, complexes and their functional attributes was entered using RACE-PRO (Rapid Annotation interfaCE for PRotein Ontology), a web-based community annotation interface for PRO (22). The RACE-PRO entries were used to create ontology terms and annotation to populate the PRO via a semiautomated process. In this way, PRO provided the ontological framework to capture the knowledge collected via text mining and data mining. The information in PRO was used to build Cytoscape protein networks displaying kinase-phosphorylated substrate relationships and PPIs. Details of each step of this workflow are provided in the following sections.
Figure 1.
Overview of the workflow for the construction of phosphorylation-focused PPI networks.
Retrieval of relevant articles about phosphorylated proteins
To identify relevant articles for creating spindle checkpoint phosphorylation networks, we used RLIMS-P version 2.0. Given the PMIDs retrieved by a PubMed search for articles containing the keywords ‘Bub1’, ‘BubR1’ or ‘Mad3’, RLIMS-P identified the subset of abstracts with phosphorylation information and generated a report page displaying (i) a table summarizing the kinase, substrate and site for each abstract; (ii) a list of suggested UniProtKB identifiers for the kinases and substrates identified; and (iii) a link to the title and abstract with the kinase, substrate and site mentions highlighted for evidence attribution. We validated the information provided by RLIMS-P, consulting the full-length article for clarification when necessary. If the abstract contained a bone fide mention of a phosphorylated protein form that had not been previously captured in PRO, we proceeded to read the full-length article to identify functional information about the phosphorylated protein and any other proteins examined in the same study.
Data mining of PPI information
To expand the BUB1B and MAD3 interaction networks, we identified additional binding partners by collecting PPI data from the following databases: MINT (Molecular INTeraction Database; release date 26 October 2012) (23); IntAct (release 159) (24); and BioGRID (Biological General Repository for Interaction Data sets; release 3.1.94) (25). We focused exclusively on physical interactions documented in low-throughput experiments, such as immunoprecipitation, affinity purification and targeted two-hybrid assays that we had not captured by text mining. We filtered out IntAct interactions that were based on spoke expansion (connecting bait to all preys in the case of a co-complex involving more than two molecules). The information about interacting partners identified by data mining was added as functional annotations including the evidence source to the BUB1B or MAD3 PRO gene product level terms.
Integration of data into the PRO
PRO ontology framework
PRO is an Open Biomedical Ontologies (OBO) Foundry ontology that provides a hierarchical representation of proteins and protein complexes (7, 26). PRO provides the ability to formally organize and integrate representations of precise protein forms so as to enhance accessibility results of protein research. PRO encompasses three subontologies: (i) ProEvo subontology, representing proteins translated from different but evolutionarily related (homologous) genes; (ii) ProForm subontology representing experimentally observed proteoforms encoded by a single gene, where ‘proteoform’ refers to the different molecular forms in which the protein product of a single gene can be found, including forms arising owing to genetic variations, alternatively spliced RNA transcripts, co-translational modification or PTM (27) and (iii) ProComp subontology representing specific amino acid chain–containing complexes. ProComp interoperates with the macromolecular complex branch of the GO cellular component subontology, as terms in this branch serve as parents to PRO complexes (28).
The PRO framework enabled the addition of functional annotation to specific protein forms and complexes. These annotations were saved in the PRO-association file (PAF) and were formulated using GO terms, including the corresponding relation.
RACE-PRO annotation interface
All the information for the various protein forms and complexes collected via text mining and data mining was entered into RACE-PRO, a web-based interface that facilitates defining and annotating protein objects without requiring knowledge of ontologies or formatting restrictions. The input to RACE-PRO is a UniProtKB identifier, a protein sequence or a PRO identifier. The RACE-PRO interface has two sections, one for defining the protein form where the user can add the name of the protein form, the protein length, the type and sites of modification, the modifying enzyme and the evidence source, and the other for functional annotation where the user can add GO terms and other vocabularies. Annotations can be accompanied by modifier words, such as ‘NOT’, which is regularly used in GO annotation, and also ‘increased’, ‘decreased’ and ‘altered’ to indicate that the properties of the protein form differ from those of a reference form (indicated in the ‘Relative to’ field). PPI information was input using the GO term ‘protein binding’ (GO: 0005515) or a child term, when appropriate; the interacting partner was entered in the ‘Interaction with’ field.
Generation of PRO terms
All RACE-PRO entries were checked by a PRO editor and then used to generate PRO terms (OBO stanzas) via a semiautomated process. In this process, the hierarchy is automatically built based on the information of the isoform or modified forms. The program first searches for existing PRO terms and generates the needed parent terms to complete the branch using information from UniProtKB. The program automatically generates standard names and definitions for the gene product and the isoform levels, whereas for the modification level, manual review is needed. PRO terms for complexes are manually created after the individual components have been entered in RACE-PRO. Annotations are automatically formatted to the PAF standard. All terms and annotations generated in this study can be found in PRO release 31.0.
Visualization of the protein networks
We used a partially automated process to display the BUB1B interaction network with Cytoscape (8). We started with a small set of human BUB1B protein forms and complexes: BUB1B (PR:000026903), its four phosphorylated forms (PR:000035362, PR:000035428, PR:000035432 andPR:000035435) and two complexes [BUB1B/BUB3 (PR:000035563) and BUB1B/BUB3/MAD2/CDC20 (PR:000035511)]. For each of these protein forms/complexes, we retrieved the PRO IDs of three types of interactors: binding partners, kinases for the phosphorylated forms and substrates for the kinases (i.e. phosphorylated forms that were phosphorylated by the protein form/complex). This information can be extracted from the PRO OBO file and PAF, which can be downloaded from the PRO Web site. Binding partners are listed in the PAF with the GO evidence code ‘inferred from physical interaction’ or IPI. Kinases are found in the comment field of the OBO stanza. Although this field is free-text, curators use a standard format to enter the information [Kinase = (‘name’; PRO ID)], thereby facilitating automated extraction. We then repeated the procedure, extracting the interactors of the interactors identified in the previous round, continuing until no new PRO IDs were found. This process resulted in a list of 73 proteins. From the PRO OBO stanzas for these proteins, we used a script to extract the following additional information: name, definition, category, label (PRO-short-label), parent–child relationship and complex components (in the case of complexes). The script then generated two tab-delimited text files, which are importable into Cytoscape: a network file containing each pair of interacting proteins, its interaction type and corresponding evidence, and a PRO entry information file containing PRO ID and entity description. Those two files were further converted into visualized protein networks with the Cytoscape functions ‘Import->Network from table’ and ‘Import->Attribute from table’. In these networks, each node is a PRO entry, and two nodes were connected by an edge if they were associated by a relation. Entity descriptions and relations annotations were represented as node or edge attributes. Scripts are available on request.
Results
Assessment of the current state of phosphorylation event curation
The scientific literature contains a wealth of protein phosphorylation data derived both from traditional low-throughput experiments that focus on a small number of proteins and from high-throughput experiments that attempt to assess the phosphorylation state of the whole proteome. This information is currently being captured in a number of high-quality manually curated resources. We aggregated the phosphorylation information from seven such databases to determine the current extent of phosphorylation event curation. Three of the databases specialize in a single organism: HPRD (human), PhosphoGrid (budding yeast) and PhosPhAt (Arabidopsis thaliana); PhosphoSitePlus, PhosphoELM and the plant database P3DB cover a small number of model organisms. UniProtKB covers a much wider range of organisms but does not specialize in the curation of phosphorylation information.
Integration of the data was done by mapping all data to UniProtKB entries. The numbers of phosphoproteins and protein kinases were determined by counting the distinct UniProtKB identifiers in the mapped data (Tables 1 and 2). As shown in Table 1, the combined databases contain curated information on >28 000 phosphoproteins, >125 000 phosphorylation sites and ∼700 protein kinases. This information is derived from >10 000 scientific publications.
Table 1.
Total phosphoproteins, phosphorylation sites, kinases and scientific publications (PMIDs) curated by seven databases that capture protein phosphorylation information (UniProtKB, Phospho.ELM, PhosphoSitePlus, HPRD, PhosphoGrid, PhosPhAt and P3DB)
| Total | |
|---|---|
| Phosphoproteins* | 28 158 |
| Sites | 125 896 |
| Kinases* | 689 |
| Sites w/ kinase information | 12 702 |
| PMIDs | 10 213 |
*The numbers of phosphoproteins and kinases are the numbers of distinct UniProtKB identifiers that were obtained by mapping the data to UniProtKB entries.
Table 2.
Number of phosphoproteins (distinct UniProtKB identifiers) for the top 15 annotated organisms
| Organism | No of phospho-proteins |
|---|---|
| H. sapiens (human) | 8738 |
| A. thaliana (mouse-ear cress) | 6423 |
| Mus musculus (mouse) | 3533 |
| Saccharomyces cerevisiae (budding yeast) | 2649 |
| Oryza sativa subsp. japonica (rice) | 2525 |
| Schizosaccharomyces pombe (fission yeast) | 969 |
| Drosophila melanogaster (fruit fly) | 750 |
| O. satvia subsp. indica (rice) | 639 |
| Caenorhabditis elegans (worm) | 583 |
| Rattus norvegicus (rat) | 555 |
| Medicago truncatula (barrel medic) | 111 |
| Bos taurus (bovine) | 87 |
| Zea mays (maize) | 86 |
| Danio rerio (zebrafish) | 86 |
| Gallus gallus (chicken) | 66 |
The number of phosphoproteins for the 15 most highly annotated organisms is shown in Table 2. This table reveals that the amount of phosphorylation information is uneven across organisms with the bulk of the data coming from humans and a few model organisms such as budding yeast. A recent report estimated the total number of human protein coding genes at ∼20 000 (29). The human phosphoproteins in the aggregated data set map to 8611 genes, or ∼43% of the total. Similarly, in budding yeast, the aggregated phosphoprotein data maps to 2458 genes, which represents ∼43% of the estimated 5300 total yeast genes (30). Based on these percentages, we would expect the number of phosphoproteins in most organisms listed in Table 2 to number in the thousands; however, there are only five organisms with >1000 curated phosphoproteins and only 11 organisms with >100.
The absence of phosphorylation information from the aggregated data set can be explained by a lack of experimental data and/or by gaps in the curation of existing data. On the experimental side, the number of characterized phosphorylated proteins is expected to grow rapidly owing to the advancement of high-throughput proteomics technologies (31, 32). To assess the degree to which curation efforts to date have captured existing phosphorylation data, we used the text-mining tool RLIMS-P to determine the total number of PubMed abstracts containing phosphorylation-related information (see ‘Materials and Methods’ section). Out of >22 million abstracts, RLIMS-P flagged ∼143 000 (0.65%) as containing references to phosphorylation. Approximately 90% of the 10 213 PMIDs in the aggregated phosphorylation data set (Table 1) were included in the article set identified by RLIMS-P, indicating that RLIMS-P has a high recall rate. In our benchmarking studies, the false-positive rate for RLIMS-P was <5% (i.e. precision >95%) (33). Thus, 143 000 is likely to be a good estimate of the number of phosphorylation-related articles currently in the literature. The 10 213 articles that have been curated so far represent only 7% of the total 143 000, indicating that much of the available phosphorylation information has yet to be captured.
In addition to the sheer amount of phosphorylation information that remains to be curated, there are also gaps in the curation of some aspects of protein phosphorylation, namely (i) representation of the multiply phosphorylated forms, (ii) capture of phosphorylation-state–specific PPIs and (iii) capture of kinase–substrate relationships.
Representation of multiply phosphorylated forms
The databases described above curate phosphorylation information on a site-by-site basis; however, proteins often exhibit multisite phosphorylation, sometimes by multiple kinases. This type of information cannot be unambiguously obtained from typical high-throughput data sets. While there has been some advancement in mass spectrometry technology for detecting multiply phosphorylated forms (34,35), most of the available information comes from single-protein studies and must be gathered by manual curation of the scientific literature. Two resources that represent the complexity of multiply phosphorylated protein forms are Reactome and PRO.
Reactome curates biological reactions and pathways, primarily in humans. Features of reaction and pathway participants, including phosphorylation site information, if known, are described in detail. Currently, Reactome contains 690 human phosphorylation reactions, including both small molecule and protein phosphorylation events. Proteoforms in Reactome are currently being imported into PRO to provide the corresponding ontological view.
The PRO ProForm subontology captures individual protein forms such as posttranslationally modified forms. In the case of phosphorylation, a separate PRO term is created for each observed phosphorylated form, which often contains combinations of phosphorylation sites documented in the literature. Currently, ∼40% of phosphoproteins in PRO with site information are phosphorylated on more than one site, suggesting that multisite phosphorylation is common and needs to be taken into account to develop a realistic picture of biological phosphorylation events.
Curation of phosphorylation-state–specific PPIs
Another area for further development is the annotation of phosphorylation-specific PPIs. Phosphorylation often plays an important role in regulating PPIs. Of the phosphorylated protein forms in PRO that have functional annotation, ∼40% are annotated with PPI information. However, major PPI resources such as IntAct, MINT and BioGRID do not focus on systematic curation of PPIs at the phosphorylation-state level. STRING, a popular web-based tool that creates and displays protein networks based on PPIs documented in many curated databases also does not incorporate phosphorylation state information when representing protein-binding events.
Curation of kinase–substrate relationships
Curation of kinase–substrate relationships is lagging behind curation of phosphorylation site information. Only ∼10% (12 702/125 896) of phosphorylation sites are associated with a specific kinase in the aggregated phosphorylation data set (Table 1). This is largely due to the fact that mass spectrometric experiments yield extensive data on phosphorylation sites but do not provide information on which kinases are responsible for the phosphorylation. Although the STRING database contains some information on kinase–substrate relationships, these relationships comprise a small fraction of the total associations. There are >9 million protein-binding interactions in STRING and only ∼73 000 interactions (0.8%) involving PTM, a category that includes kinase–substrate relationships.
In the sections below, we use the spindle checkpoint as a case study to illustrate a curation workflow and network generation procedure that addresses these deficiencies in phosphorylation event curation. We focus on the capture of phosphorylated forms, including those phosphorylated on multiple sites, phosphorylation-specific PPIs and kinase–substrate relationships.
Analysis of the spindle checkpoint using text and data mining
We conducted our initial analysis on the core spindle checkpoint proteins BUB1, BUB1B and MAD3. BUB1 is highly conserved from yeast to humans. MAD3 is the closest yeast relative of human BUB1B. The N-terminal half of BUB1B shares homology with MAD3, but BUB1B contains a C-terminal kinase-like domain that is absent from MAD3 (36). PubMed searches for ‘Bub1’, ‘BubR1’ and ‘Mad3’ returned 583, 371 and 121 articles, respectively, from which RLIMS-P identified 88 (15%) of the ‘Bub1’ abstracts, 69 (19%) of the ‘BubR1’ abstracts and 16 (13%) of the ‘Mad3’ abstracts as containing mentions of kinase, substrate and/or phosphorylation site. The rate of RLIMS-P positive articles was significantly higher for these three proteins than for PubMed as a whole, in which ∼0.65% were flagged by RLIMS-P. This indicates that research on BUB1, BUB1B and MAD3 is enriched for studies of phosphorylation and points to the importance of phosphorylation in the spindle checkpoint response. Because BUB1, BUB1B and MAD3 have closely related functions, they are often discussed together in the same article. When this overlap is taken into account, the number of unique abstracts identified by RLIMS-P was 120.
We curated the full text of 68 of the 120 articles. Note that the 52 articles that we chose not to pursue further were not false positives. Most of them fell into one of two categories: articles that contained a brief mention of a phosphorylated form that had been discussed in depth in an article that we had already processed, or articles that did not contain new experimental data (e.g. review articles). Interestingly, some of the articles identified by RLIMS-P described phosphorylation events that did not involve BUB1, BUB1B or MAD3 as kinase or substrate (i.e. the mention of BUB1/BUB1B/MAD3 and the mention of the phosphorylation event were not directly related). However, because we were interested in the spindle checkpoint as a whole, not just in the individual BUB1, BUB1B and MAD3 proteins, we extracted information from these articles as well. In total, we created RACE-PRO entries for 182 proteoforms, including 71 phosphorylated forms. Even though our ‘Bub1’ search returned the highest number of PubMed and RLIMS-P results, we found that the articles we curated contained by far the most detailed information on BUB1B and its phosphorylated forms. Therefore, we decided to center the rest of our analysis on human BUB1B and its yeast homolog MAD3.
To capture additional PPI information, we expanded our analysis to include three curated PPI databases—MINT, IntAct and BioGrid. After excluding interactions that were seen only in high-throughput experiments and interactions that were likely to be indirect, we found 17 BUB1B binding proteins and three MAD3 binding proteins that we had not previously identified through text mining. These interactions were added to the annotation of the gene level BUB1B and MAD3 terms. (In PRO, a gene level term is one that encompasses all proteoforms of a given protein-coding gene.)
The human BUB1B network
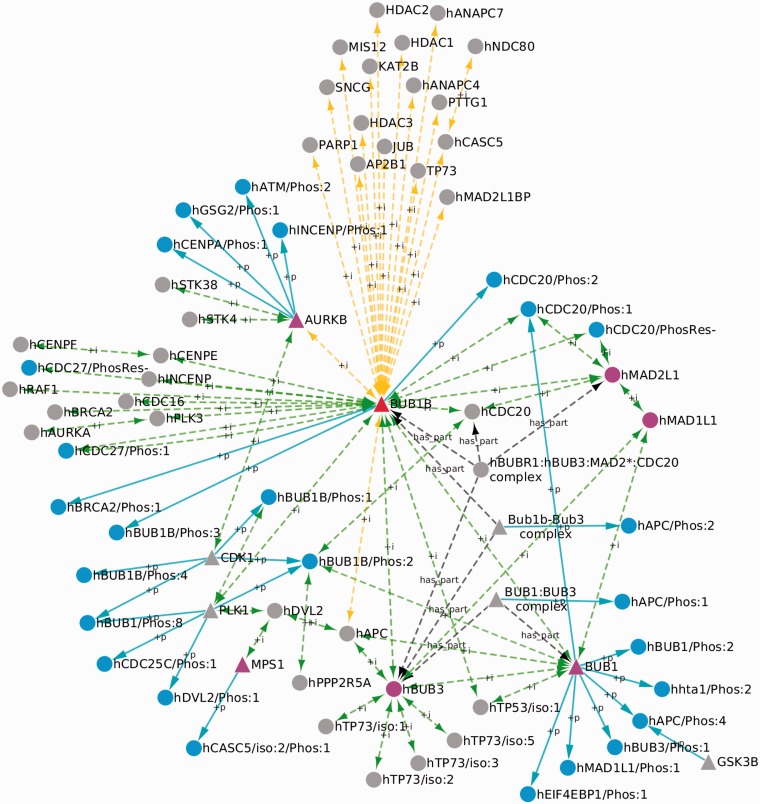
The BUB1B protein interaction network based on our text and data mining results is shown in Figure 2. Five of the seven core checkpoint proteins—MAD1L1, MAD2L1, BUB1, BUB3 and MPS1 (Figure 2, purple nodes)—are linked directly or indirectly to BUB1B via interactions we identified by text mining. AURKB, on the other hand, was incorporated into the network through a physical interaction with BUB1B that we identified by data mining. Thus, both text and data mining contributed critical pieces to the spindle checkpoint network.
Figure 2.
The human BUB1B network. The BUB1B node is shown in red; other core spindle checkpoint proteins are shown in purple; nodes representing phosphorylation-state–specific forms are shown in blue. Triangles indicate protein kinases. Green and yellow edges are PPIs identified by text mining and data mining, respectively; blue edges connect kinases to their phosphorylated products; black edges indicate the has_part relation connecting protein complexes to their components.
The prominent role that phosphorylation plays in the checkpoint is evident from our network. The network consists of 73 nodes, 26 of which have a specified phosphorylation state: 24 are phosphorylated forms and two are unphosphorylated forms (Figure 2, blue nodes). PRO terms are created for unphosphorylated protein forms that have been experimentally characterized (e.g. through the use of phosphorylation site mutants or kinase inhibitors). There are 26 kinase–substrate relationships (Figure 2, blue arrows) involving nine protein kinase nodes (BUB1B, the BUB1B/BUB3 complex, BUB1, the BUB1/BUB3 complex, CDK1, PLK1, GSK3B, MPS1 and AURKB; Figure 2, triangular nodes). The number of kinase–substrate relationships is greater than the number of phosphorylated forms because in some cases multiple kinases contribute to the formation of a single phosphorylated form (e.g. BUB1B/Phos:2).
Our framework also represents phosphorylation-state–specific PPIs. The BUB1B network contains 10 such interactions, which are listed in Table 3. In some cases, the interaction is specific for the phosphorylated protein form, suggesting that phosphorylation may be important for regulating the interaction. For example, one of the phosphorylated forms of BUB1B, BUB1B/Phos:1, has a phosphorylation-dependendent interaction with PLK1. In other cases, both the phosphorylated and unphosphorylated forms of the protein participate in the same PPI, indicating that phosphorylation is dispensable for the interaction. For example, both phosphorylated CDC27 (CDC27/Phos:1) and unphosphorylated CDC27 (CDC27/PhosRes-) interact with BUB1B. Both scenarios provide insight into the role of phosphorylation in the spindle checkpoint.
Table 3.
Phosphorylation-state–specific PPIs in the human BUB1B network
| Protein Form #1 | Protein Form #2 |
|---|---|
| BUB1B/Phos:1 | PLK1 |
| BUB1B/Phos:2 | BUB1 |
| BUB1B/Phos:2 | CDC20 |
| BUB1B/Phos:2 | PPP2R5A |
| CDC27/Phos:1 | BUB1B |
| CDC27/PhosRes- | BUB1B |
| CDC20/Phos:1 | BUB1B |
| CDC20/Phos:1 | MAD2L1 |
| CDC20/PhosRes- | BUB1B |
| CDC20/PhosRes- | MAD2L1 |
Much of the PPI data we captured through both text and data mining did not specify the phosphorylation state of the interacting partners. For this reason, our network contains many PPIs involving gene level protein forms. Even though BUB1B has four phosphorylated forms and several documented phosphorylation-state–specific PPIs, it also has 30 PPIs (13 captured from the scientific literature and 17 from PPI databases) that map to the gene level BUB1B node. As more experimental data on BUB1B phosphorylation and PPIs accumulates, these interactions could be remapped to more specific BUB1B forms. In the meantime, this network could be used to guide a systematic experimental inquiry into which BUB1B PPIs are affected by the BUB1B phosphorylation state.
Comparison with the BUB1B network generated by STRING
We were interested in comparing our BUB1B phosphorylation network with the BUB1B PPI network generated by STRING. As described above, STRING builds PPI networks from information contained in a number of curated resources. STRING networks can also be expanded to include predicted associations derived from sources such as gene neighborhood and gene co-expression analyses. Because it draws on such a large set of underlying data, the top-ranked interactions in STRING networks are supported by multiple lines of evidence and are highly reliable. STRING networks are based on ‘functional interactions’ between proteins and include physical and genetic interactions as well as interactions-based enzyme–substrate relationships or involvement in a common pathway. However, STRING has relatively little information on PTM and does not represent phosphorylated protein forms, so we expected that the STRING network would differ significantly from our network.
Using the STRING web interface, we constructed a BUB1B-centered network based on experimentally observed interactions. The network contained 163 BUB1B-interacting proteins with confidence scores ranging from 0.204 to 0.999. Eighty-three percent (135/163) of the interactors had a confidence score of at least 0.800. According to the STRING classification of interactions, five of the interactions—with CDC20, PLK1, AURKB, CDK1 and BRCA2—involve some type of PTM.
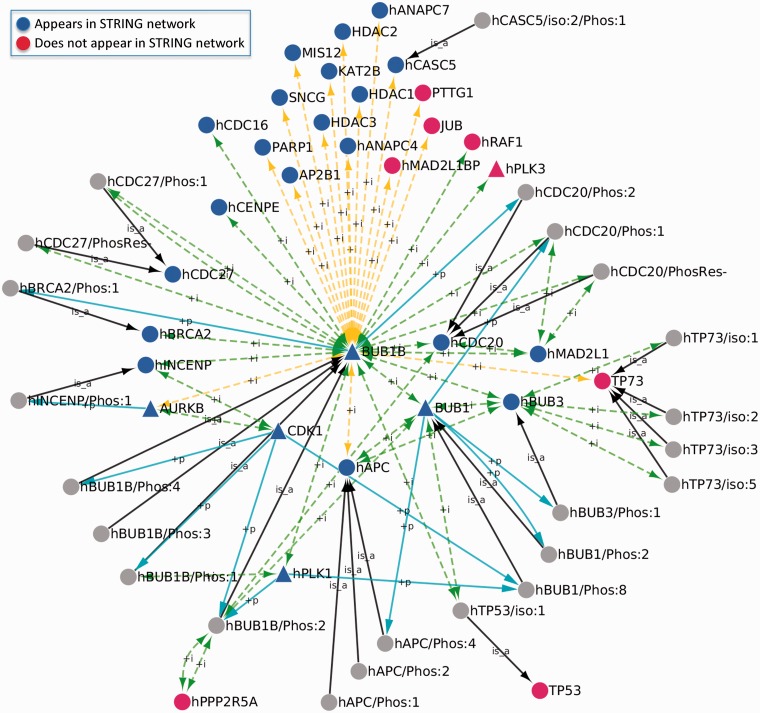
Our original network contains both direct (i.e. first neighbor) and indirect (i.e. second neighbor, third neighbor, etc.) BUB1B interactors. Because the STRING network we constructed contains only direct BUB1B interactors, we compared it with a subset of our original network that consists of direct BUB1B interactors, their isoforms and their PTM forms (Figure 3). In our subnetwork, there were 38 protein forms that interacted directly with BUB1B and/or its phosphorylated forms. The 38 protein forms were derived from 32 gene level terms. Of these 32 gene level terms, 24 (75%) appear in the STRING network (Figure 3, blue nodes), with confidence scores ranging from 0.619 to 0.999. The interactors in our network that do not appear in the STRING network (Figure 3, red nodes) were captured by data mining (AJUBA, MAD2L1BP, PTTG1 and RAF1), text mining (PLK3, PPP2R5A and p53) or both (p73). In addition, our subnetwork contains four of the five PTM interactions identified by STRING—PLK1 and CDK1 as BUB1B kinases and CDC20 and BRCA2 as BUB1B substrates. Unlike STRING, our subnetwork does not indicate a PTM between BUB1B and AURKB; instead, those two proteins are linked only by a protein-binding relationship.
Figure 3.
Comparison of the human BUB1B phosphorylation network with the human BUB1B STRING network. The portion of the BUB1B phosphorylation network comprising proteins that directly interact with BUB1B, their isoforms and their PTM forms is shown. Gene level terms that appear in the STRING BUB1B network are shown in blue; gene level terms that do not appear in the STRING network are shown in red; isoforms or PTM forms are shown in gray. Triangles indicate protein kinases. Green, yellow and blue edges are as described in Figure 2; black edges indicate the is_a relationship connecting isoforms and PTM forms to their parent gene-level forms.
Where our network departs sharply from the STRING, BUB1B network is in its granularity. In addition to BUB1B, which has four phosphorylated forms, 10 of the 32 BUB1B interacting proteins have at least one isoform or PTM form in our network. Altogether there are 23 isoforms or PTM forms in our network derived from either BUB1B or one of its direct interacting proteins. (These protein forms are connected to their parent forms by ‘is_a’ relations (black arrows) in Figure 3.) Eleven of these 23 forms (48%) participate in form-specific binding interactions with other proteins in the subnetwork (Figure 3).
Our subnetwork also has a much higher proportion of protein kinases as compared with the STRING network. Out of 33 gene level terms in our subnetwork (BUB1B and 32 BUB1B interactors), six (18%) are protein kinases (Figure 3, triangular nodes). Even though it is based on a much larger set of underlying data and contains almost five times as many proteins (BUB1B + 163 BUB1B interactors), the STRING network contains only seven protein kinases (4%). Five kinases (BUB1B, BUB1, AURKB, PLK1 and CDK1) appear in both networks, two (NEK2 and TAOK1) appear in the STRING network only and one (PLK3) appears in our subnetwork only. NEK2 and TAOK1 are included in the STRING network because they are found along with BUB1B in large protein complexes curated by Reactome. NEK2, BUB1B and 15 other proteins comprise the NEK2:MCC:APC/C complex (Reactome ID: REACT_7992.1), whereas TAOK1, BUB1B and 67 other proteins comprise the kinetochore complex (Reactome ID: REACT_14970.1). Thus, the interactions of NEK2 and TAOK1 with BUB1B are likely to be indirect and independent of NEK2 and TAOK1 kinase activity. Moreover, because Reactome data are currently being incorporated into PRO, PRO representations of these complexes will soon be available.
In summary, our example demonstrates that our curation and network building workflow can be used to provide a level of detail about protein forms and kinases that goes beyond the information available in STRING.
Comparison of the representations of BUB1B phosphorylation in PRO, Phospho.ELM and PhosphoSitePlus
As a measure of the extent to which our curation method captured phosphorylation site information from the scientific literature, we compared the information on human BUB1B phosphorylation in three databases from our aggregated data set—Phospho.ELM, PhosphoSitePlus and HPRD—to the data we captured in PRO (Table 4). Twelve BUB1B phosphorylation sites observed in low-throughput experiments are present in Phospho.ELM and/or PhosphoSitePlus. There was no low-throughput site information in HPRD. The three databases also contained 19 sites identified in high-throughput experiments only. Our text mining efforts captured 11 of the 12 low-throughput sites (92%); in addition, we uncovered one site (Thr-608) that only had high-throughput evidence in the phosphorylation databases. In PRO, we organized this information into 4 human BUB1B forms phosphorylated on different experimentally observed combinations of the 13 sites. BUB1B/Phos:1 is phosphorylated on Thr-620; BUB1B/Phos:2 is phosphorylated on Thr-620, Ser-676, Thr-680, Thr-792 and Thr-1008; BUB1B/Phos:3 is phosphorylated on Thr-608; and BUB1B/Phos:4 is phosphorylated on Ser-435, Ser-543, Ser-574, Ser-670, Ser-720 and Ser-1043.
Table 4.
BUB1B phosphorylation sites in Phospho.ELM, PhosphoSitePlus, HPRD and PRO identified in low-throughput experiments
| Site | Phospho.ELM | PhosphoSitePlus | HPRD | PRO |
|---|---|---|---|---|
| Ser-435 | LTP, HTP | LTP, HTP | HTP | BUB1B/Phos:4 |
| Ser-543 | LTP, HTP | LTP, HTP | HTP | BUB1B/Phos:4 |
| Ser-574 | HTP | LTP, HTP | BUB1B/Phos:4 | |
| Thr-608 | HTP | BUB1B/Phos:3 | ||
| Thr-620 | LTP | LTP, HTP | BUB1B/Phos:1, BUB1B/Phos:2 | |
| Ser-670 | LTP, HTP | LTP, HTP | HTP | BUB1B/Phos:4 |
| Ser-676 | LTP | LTP, HTP | BUB1B/Phos:2 | |
| Thr-680 | LTP | BUB1B/Phos:2 | ||
| Ser-720 | HTP | LTP, HTP | BUB1B/Phos:4 | |
| Thr-792 | LTP | BUB1B/Phos:2 | ||
| Ser-884 | LTP, HTP | |||
| Thr-1008 | LTP | BUB1B/Phos:2 | ||
| Ser-1043 | LTP, HTP | LTP, HTP | HTP | BUB1B/Phos:4 |
LTP, low throughput; HTP, high throughput.
In addition to the sites listed above, the following phosphorylation sites were identified in high-throughput experiments only:
PhosphoSitePlus: Ser-39, Thr-40, Thr-54, Ser-83, Thr-315, Thr-368, Ser-384, Tyr-404, Thr-471, Thr-600, Ser-633, Tyr-660, Tyr-766, Ser-797
PhosphoSite Plus and Phospho.ELM: Ser-367, Ser-537, Ser-733
PhosphoSitePlus and HPRD: Thr-434, Thr-1042.
Overall, our method captured almost all of the human BUB1B phosphorylation sites present in other phosphorylation site databases with the added advantage that we could define BUB1B multiply phosphorylated forms carrying biologically relevant combinations of sites.
Comparison of the budding yeast MAD3 and BUB1B networks
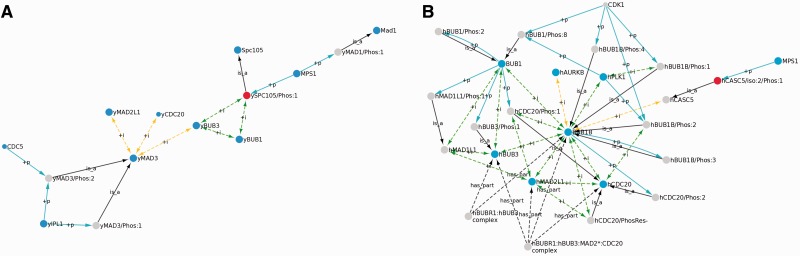
MAD3 is the closest budding yeast relative of human BUB1B. Its sequence is highly similar to the N-terminal half of BUB1B, and like BUB1B, MAD3 is essential for the spindle checkpoint response. However, MAD3 is a shorter protein than BUB1B (515 vs. 1050 amino acids) and lacks the kinase-like domain found in the C-terminal half of BUB1B (5). Although some experiments have suggested that BUB1B can phosphorylate several proteins including CDC20 (Figure 2), the role of these phosphorylation events in the spindle checkpoint is unclear (37). Moreover, it was recently shown that BUB1B lacks some critical residues conserved in most protein kinases and may instead be a pseudokinase (5). Most other components of the spindle checkpoint are well conserved in budding yeast and humans. The MAD3 network based on literature mining using RLIMS-P and data mining for additional PPIs is shown in Figure 4A.
Figure 4.
Comparison of the yeast MAD3 and human BUB1B networks. (A) The MAD3 network. (B) The portion of the BUB1B network containing the human homologs of the yeast network proteins. In (A) and (B), nodes representing homologous gene-level protein forms in yeast and humans are colored blue; nodes representing homologous phosphorylated protein forms are colored red. Edges are color-coded as in Figure 3.
The budding yeast network is much smaller than the human network because the articles identified by RLIMS-P contained relatively little information about MAD3. Many articles that mentioned MAD3 referred to it only briefly as the homolog of BUB1B. The subset of the BUB1B network that overlaps with the budding yeast network is shown in Figure 4B. All of the gene level protein forms present in the budding yeast network, including CDC20, MAD2L1, BUB1, BUB3, MAD1 (MAD1L1 homolog) CDC5 (PLK1 homolog) and IPL1 (AURKB homolog) are also present in the human network (blue nodes in Figures 4A and B). Many of the PPIs are also conserved. Budding yeast MAD3 binds to MAD2L1, BUB3 and CDC20, the same proteins that comprise the MCC in humans. In addition, the interaction of BUB1 and BUB3 is found in both networks.
In contrast to the high level of conservation of proteins and PPIs, the conservation of phosphorylated forms between human and budding yeast was low. Although MAD3 has two phosphorylated forms, neither corresponds to any of the four BUB1B phosphorylated forms. All of the BUB1B phosphorylation sites lie in the C-terminal half of the BUB1B sequence, which is not present in MAD3. Yeast MAD1 is phosphorylated by MPS1 under conditions that activate the spindle checkpoint (38), but this phosphorylation has not been observed in humans. Instead, human MAD1L1 is phosphorylated by BUB1. Finally, the phosphorylated forms of CDC20, BUB1 and BUB3 that have been observed in humans have not been observed in yeast.
Interestingly, both CASC5/iso:2/Phos:1 and its closest budding yeast relative, SPC105/Phos:1 (red nodes in Figure 4A and B), are phosphorylated by the same kinase, MPS1, and the phosphorylated forms perform the same function: recruitment of BUB1 and BUB3 to the kinetochore, an essential early step in the checkpoint response (39,40). Phosphorylation of both proteins takes place on sequences known as MELT motifs, but in SPC105 these motifs are located in the N-terminus of the protein, whereas in CASC5 they are located in the middle of the protein. Thus, phosphorylation is carried out by the same kinase on the same motif and has the same functional consequences, yet the phosphorylated motif is located in different regions of the two proteins.
Discussion
We have described a representation of the role of phosphorylation for a few core proteins in the spindle checkpoint, which we developed using a combination of text mining, data mining, ontologies and network visualization tools. Our approach addressed some areas that have been neglected by other curation methods, but also highlighted some of the challenges inherent in phosphorylation event curation.
Capture of spindle checkpoint phosphorylation knowledge
By identifying articles with mentions of phosphorylation, RLIMS-P allows curators to focus on a relatively small number of highly relevant documents in a long list of PubMed search results. For the three proteins we studied here (BUB1, BUB1B and MAD3), only ∼15% of the total documents identified by PubMed were flagged by RLIMS-P. The set of 120 articles identified by RLIMS-P was a manageable number for in-depth curation of the full-length text. Through text mining, we identified 11 of the 12 human BUB1B phosphorylation sites that were documented with low-throughput evidence in our aggregated phosphorylation site data set. The site that we did not capture, Ser-884, was described in the full text of an article, but not in the abstract, so it was not detected by RLIMS-P. An expanded version of RLIMS-P that searches for phosphorylation-related information in full-length articles is currently being developed. Moreover, we identified low-throughput evidence for Thr-608 phosphorylation of BUB1B, whereas the phosphorylation databases had only high-throughput evidence for this site. We also captured all but one of the PTM relationships present in the STRING BUB1B network even though our network contained a much smaller number of BUB1B interactors (32 as opposed to 163). The significance of the one STRING PTM relationship—between BUB1B and AURKB—that did not appear in our network is unclear. We were unable to determine the evidence for this interaction from the STRING Web site. There was no record of BUB1B phosphorylating AURKB or vice versa in the seven phosphorylation resources we used for our aggregate analysis, nor were we able to find text evidence for a direct kinase–substrate relationship between BUB1B and AURKB using either a BUB1B-centric or AURKB-centric RLIMS-P search. One possibility is that the interaction involves a PTM other than phosphorylation, as STRING does not specify the type of PTM. Overall, this work demonstrates that our method, when applied to a particular biological process, is capable of providing a comprehensive picture of the role of phosphorylation in that process.
The majority of phosphorylation site information is currently being generated by medium-scale or large-scale mass spectrometry experiments that collect phosphorylation data on many proteins simultaneously. This type of data poses two challenges for our curation strategy. First, individual proteins of interest are unlikely to be mentioned in the abstracts of publications describing the phosphorylation of many proteins; instead this information is often recorded in supplementary tables that are not accessible to PubMed or RLIMS-P searches. The second problem is that high-throughput mass spectrometric experiments typically analyze phosphorylation of peptide fragments of proteins, so they do not provide information on phosphorylation site combinatorics. Thus, one of the unique features of our approach—the definition of full-length phosphorylated protein forms, including multiply phosphorylated forms—is not compatible with data from these experiments. Top-down proteomics strategies, which can determine the modification status of large protein fragments or even entire proteins, and hybrid approaches that combine mass spectrometric analyses of whole proteins and protein fragments, are a promising sources of phosphorylation combinatorics data (34,35). We are currently engaged in incorporating information derived from these approaches into PRO.
Phosphorylation specific PPIs
One of the major innovations of our approach is the inclusion of phosphorylation state information in PPI networks. Our BUB1B network contains 10 such relations, and we are working to increase that number. All of the information on phosphorylation-state–specific PPIs must come from literature curation as PPI databases do not systematically curate phosphorylation information. Extracting this information requires time-consuming manual curation. While this process is already made easier by the use of RLIMS-P, which prioritizes articles containing phosphorylation information, we plan to streamline it further with the introduction of the eFIP (extracting functional impact of phosphorylation) text-mining tool (41) into our curation pipeline. Given a gene or protein name as input, eFIP flags relevant abstracts that refer to PPIs involving phosphorylated proteins. A full-scale eFIP processing of the ∼143 000 RLIMS-P-positive articles in PubMed resulted in the identification of ∼10 000 (7%) articles describing phospho-specific PPIs. The use of eFIP will allow us to focus curation efforts on articles that are most pertinent to construction of phosphorylation-centric PPI networks.
Evidence quality
While conducting this case study, we encountered variability in the quality of the evidence used to support assertions. For example, phosphorylation-site information was sometimes based on in vivo experiments, which are likely to be biologically relevant, and sometimes on in vitro experiments, whose relevance is less clear. In addition, we found examples of outright disagreement in the literature, such as over the question of whether or not BUB1B has kinase activity (5,42). Currently, in PRO, the degree of confidence in the content is indicated by the wording of PRO term definitions, by the addition of free text comments to PRO terms and by the provision of links to the source data (usually a PubMed ID). To express the uncertainty regarding BUB1B kinase activity, for example, the PRO term for the putative autophosphorylated form of BUB1B, BUB1B/Phos:3 (PR:000035431), contains the comment ‘There is some controversy in the literature on whether BUB1B has kinase activity or is a pseudokinase.’ This solution is not ideal because it is highly subjective, difficult to apply consistently and can only be interpreted by a human reader. This issue could be addressed through the use of machine-readable evidence codes similar to those used by GO. These codes could also be used to add confidence information to our PPI network displays.
Applications of phosphorylation networks
Our phosphorylation networks are valuable for the cross-species analysis of proteins, PPIs and phosphorylated forms in a biological process. In our case study, we found that spindle checkpoint proteins and gene level PPIs were conserved in budding yeast and humans, but phosphorylated forms were not. These results suggest that events (such as PPIs) that depend on protein phosphorylation state in humans may not occur in budding yeast. For example, the phosphorylation-dependent interaction of BUB1B/Phos:2 with the protein phosphatase 2A subunit, PPP25A, appears to be important for regulating microtubule-kinetochore interactions, a function which so far has not been attributed to MAD3. Based on our analysis, we would predict that MAD3 would not interact with protein phosphatase 2A in budding yeast; it would be interesting to experimentally test this prediction. Human phosphorylated forms are more likely to be conserved in closely related species, such as other mammals. For example, all of the phosphorylation sites in the four human BUB1B phosphorylated forms are conserved in mouse and rat and several of the sites have been experimentally shown to be phosphorylated in mouse (43).
Finally, the phosphorylation network provides a guide to the identification of new phospho-specific PPIs. Proteins in the network that have phosphorylated forms and gene level PPIs could be tested to see if any of its PPIs are affected by phosphorylation state.
Conclusions
Our integrated approach enhances existing phosphorylation event representation by providing a framework for the definition and annotation of biologically relevant phosphorylated forms, including multiply phosphorylated forms. Through its display of phosphorylation-specific PPIs, it brings together phosphorylation and PPI information that is usually curated separately. Annotation is done in a machine-readable format that allows for the semiautomated display of PPI networks. Our approach can be applied to any biological process that involves phosphorylation. The curation process uses user-friendly publicly available tools, including RLIMS-P and RACE-PRO, and we invite community participation in the development of phosphorylation networks for their biological processes of interest.
Acknowledgements
The authors would like to thank Peter D'Eustachio for assistance with searching Reactome and Natalia Roberts for providing phosphorylation site information from PRO.
Funding
This work has been supported by the National Science Foundation [ABI-1062520] and the National Institutes of Health [2R01GM080646-06 and 3R01GM80646-07S1]. Funding for open access charge: National Institutes of Health [3R01GM80646-07S1].
Conflict of interest. None declared.
References
- 1.Kim S, Yu H. Mutual regulation between the spindle checkpoint and APC/C. Semin. Cell Dev. Biol. 2011;22:551–558. doi: 10.1016/j.semcdb.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 3.Zich J, Hardwick KG. Getting down to the phosphorylated ‘nuts and bolts' of spindle checkpoint signalling. Trends Biochem. Sci. 2010;35:18–27. doi: 10.1016/j.tibs.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Welburn JP, Vleugel M, Liu D, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suijkerbuijk SJ, van Dam TJ, Karagoz GE, et al. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Oh HJ, Kim MJ, Song SJ, et al. MST1 limits the kinase activity of aurora B to promote stable kinetochore-microtubule attachment. Curr. Biol. 2010;20:416–422. doi: 10.1016/j.cub.2009.12.054. [DOI] [PubMed] [Google Scholar]
- 7.Natale DA, Arighi CN, Barker WC, et al. The Protein Ontology: a structured representation of protein forms and complexes. Nucleic Acids Res. 2011;39:D539–D545. doi: 10.1093/nar/gkq907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinkel H, Chica C, Via A, et al. Phospho.ELM: a database of phosphorylation sites—update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornbeck PV, Kornhauser JM, Tkachev S, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark C, Su TC, Breitkreutz A, et al. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database. 2010;10:28. doi: 10.1093/database/bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao Q, Bollinger C, Gao J, et al. P(3)DB: an integrated database for plant protein phosphorylation. Front. Plant Sci. 2012;3:206. doi: 10.3389/fpls.2012.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durek P, Schmidt R, Heazlewood JL, et al. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zulawski M, Braginets R, Schulze WX. PhosPhAt goes kinases—searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res. 2013;41:D1176–D1184. doi: 10.1093/nar/gks1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011 doi: 10.1093/database/bar009. doi:10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan X, Hu Z, Wu H, et al. An online literature mining tool for protein phosphorylation. Bioinformatics. 2006;22:1668–1669. doi: 10.1093/bioinformatics/btl159. [DOI] [PubMed] [Google Scholar]
- 18.Croft D, O'Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–S697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LJ, Kuhn M, Stark M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–S416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu ZZ, Narayanaswamy M, Ravikumar KE, et al. Literature mining and database annotation of protein phosphorylation using a rule-based system. Bioinformatics. 2005;21:2759–2765. doi: 10.1093/bioinformatics/bti390. [DOI] [PubMed] [Google Scholar]
- 22.Arighi CN. A tutorial on protein ontology resources for proteomic studies. Methods Mol. Biol. 2011;694:77–90. doi: 10.1007/978-1-60761-977-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatr-aryamontri A, Ceol A, Palazzi LM, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerrien S, Aranda B, Breuza L, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceusters W, Smith B. A unified framework for biomedical terminologies and ontologies. Stud. Health Technol. Inform. 2010;160:1050–1054. [PMC free article] [PubMed] [Google Scholar]
- 27.Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bult C, Drabkin H, Evsikov A, et al. The Representation of Protein Complexes in the Protein Ontology (PRO) BMC Bioinformatics. 2011;12:371. doi: 10.1186/1471-2105-12-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackiewicz P, Kowalczuk M, Mackiewicz D, et al. How many protein-coding genes are there in the Saccharomyces cerevisiae genome? Yeast. 2002;19:619–629. doi: 10.1002/yea.865. [DOI] [PubMed] [Google Scholar]
- 31.Pan J, Borchers CH. Top-down structural analysis of posttranslationally modified proteins by Fourier transform ion cyclotron resonance-MS with hydrogen/deuterium exchange and electron capture dissociation. Proteomics. 2013;13:974–981. doi: 10.1002/pmic.201200246. [DOI] [PubMed] [Google Scholar]
- 32.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 33.Narayanaswamy M, Ravikumar KE, Vijay-Shanker K. Beyond the clause: extraction of phosphorylation information from medline abstracts. Bioinformatics. 2005;21:i319–i327. doi: 10.1093/bioinformatics/bti1011. [DOI] [PubMed] [Google Scholar]
- 34.Tran JC, Zamdborg L, Ahlf DR, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prabakaran S, Everley RA, Landrieu I, et al. Comparative analysis of Erk phosphorylation suggests a mixed strategy for measuring phospho-form distributions. Mol. Syst. Biol. 2011;7:15. doi: 10.1038/msb.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elowe S. Bub1 and BubR1: at the Interface between chromosome attachment and the spindle checkpoint. Mol. Cell. Biol. 2011;31:3085–3093. doi: 10.1128/MCB.05326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardwick KG, Weiss E, Luca FC, et al. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 39.Yamagishi Y, Yang CH, Tanno Y, et al. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell. Biol. 2012;14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 40.London N, Ceto S, Ranish JA, et al. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tudor CO, Arighi CN, Wang Q, et al. The eFIP system for text mining of protein interaction networks of phosphorylated proteins. Database. 2012;5:bas044. doi: 10.1093/database/bas044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Y, Kim C, Ahmad S, et al. CENP-E–dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J. Cell Biol. 2012;198:205–217. doi: 10.1083/jcb.201202152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegemann B, Hutchins JR, Hudecz O, et al. Systematic phosphorylation analysis of human mitotic protein complexes. Sci. Signal. 2011;4:2001993. doi: 10.1126/scisignal.2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]