Fig 2.
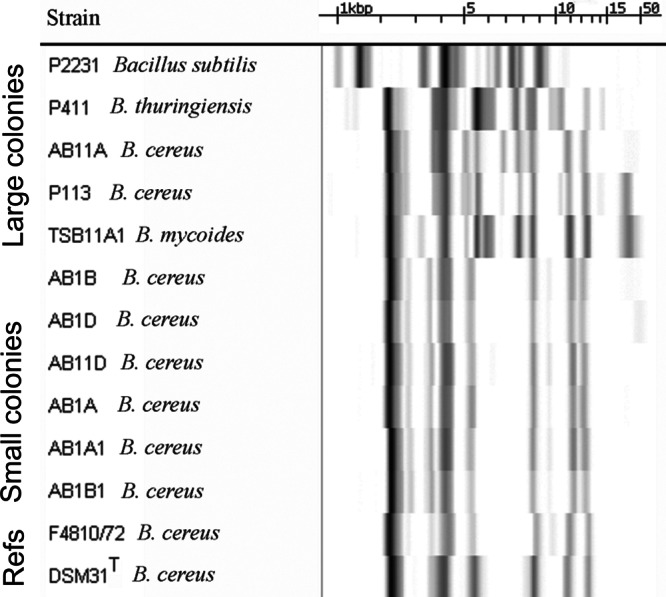
Fingerprints of the ribosomal operon area of Bacillus sp. isolated from potato tubers. Ribopatterns were generated with the automated Riboprinter using restriction enzyme EcoRI and visualized by hybridization to phosphorescence-labeled ribosomal operon of E. coli. The figure shows the identical ribopatterns of cereulide-producing, small colonies (Fig. 1), in contrast to the diversity of ribopatterns of the large-colony-forming B. cereus sensu lato that did not produce cereulide. The species names are indicated by the nearest matching ribopattern using a commercial library and an in-house database (120 well-characterized strains) of B. cereus. Patterns of strains B. cereus DSM31T (ATCC 14579T) and F4810/72 are shown as a reference. The measure bar shows the calibration of the fragment sizes in kilobases, obtained with molecular marker mixes of 1.1, 2.2, 3.2, 6.5, 9.6, and 48 kb.
