Fig 1.
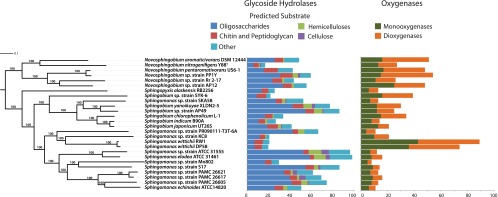
Maximum-likelihood MLSA of 26 sphingomonad genomes constructed from concatenated nucleotide sequences of 268 universally conserved genes, together with the distribution of glycoside hydrolases (GHs) and mono- and dioxygenases identified in the sphingomonad genomes. The topology of this tree is identical to the AAI dendrogram created from the proteins encoded in these genomes. Bootstrap support for the MLSA is given at each node, and the bar indicates the number of nucleotide substitutions per site. GHs were divided into categories based on their predicted substrate as follows: oligosaccharides, GH1, GH2, GH3, GH13, GH15, GH28, GH29, GH31, GH35, GH36, GH39, GH42, GH43, and GH92; hemicelluloses, GH8, GH10, GH26, GH51, GH53, and GH105; chitin and peptidoglycan, GH18, GH20, GH23, GH24, GH25, and GH73; cellulose, GH5, GH6, GH9, and GH44; other, all other GH families.
