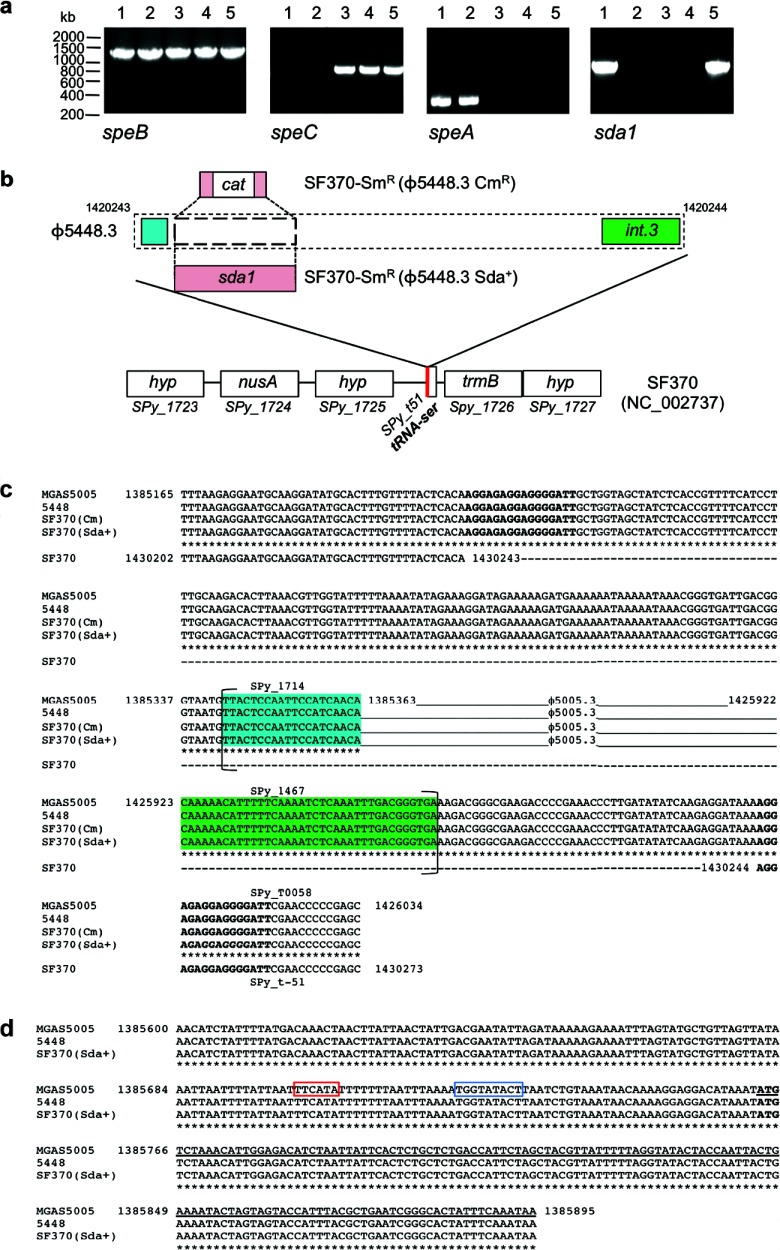
Fig 2.
Genetic characterization of SF370-Smr lysogens. (a) PCR amplification of GAS virulence genes speB, speC, speA, and sda1. Lanes 1, 5448; lanes 2, 5448Δsda; lanes 3, SF370-Smr; lanes 4, SF370-Smr(ϕ5448.3 Cmr); lanes 5, SF370-Smr(ϕ5448.3 Sda+). (b) Schematic illustrating the M1T1 ϕ5448.3 phage insertion site within the M1 SF370 genome backbone. In SF370-Smr(ϕ5448.3 Cmr), the chloramphenicol resistance gene replaces the sda1 open reading frame of phage ϕ5448.3. The red line indicates the phage insertion point immediately adjacent to a tRNA-Ser sequence. (c) Alignment of the Sda1-encoding bacteriophage attachment sites in wild-type M1T1 strains (MGAS5005 and 5448) and M1 lysogens SF370-Smr(ϕ5448.3 Cmr) and SF370-Smr(ϕ5448.3 Sda+). The sequences of the wild-type strains and the lysogens are identical. Phage insertion causes duplication of the sequence at the insertion point, resulting in a set of direct repeats (boldface). Brackets indicate the boundaries of open reading frames. The highlighted sequences represent the first (blue) and last (green) open reading frames of phages ϕ5005.3 and 5448.3. (d) Alignment of the insertion site of the sda1 gene within the Sda1-encoding bacteriophage in wild-type M1T1 strains (MGAS5005 and 5448) and the M1 lysogen SF370-Smr(ϕ5448.3 Sda+). The sequences are identical. sda1 insertion by double crossover has occurred without disruption of the sda1 open reading frame or of the 5′ sequence containing the promoter region (the −35 box is framed in red and the −10 box in blue). The starting codon for the sda1 gene is shown in boldface, and the sda1 open reading frame is underlined.