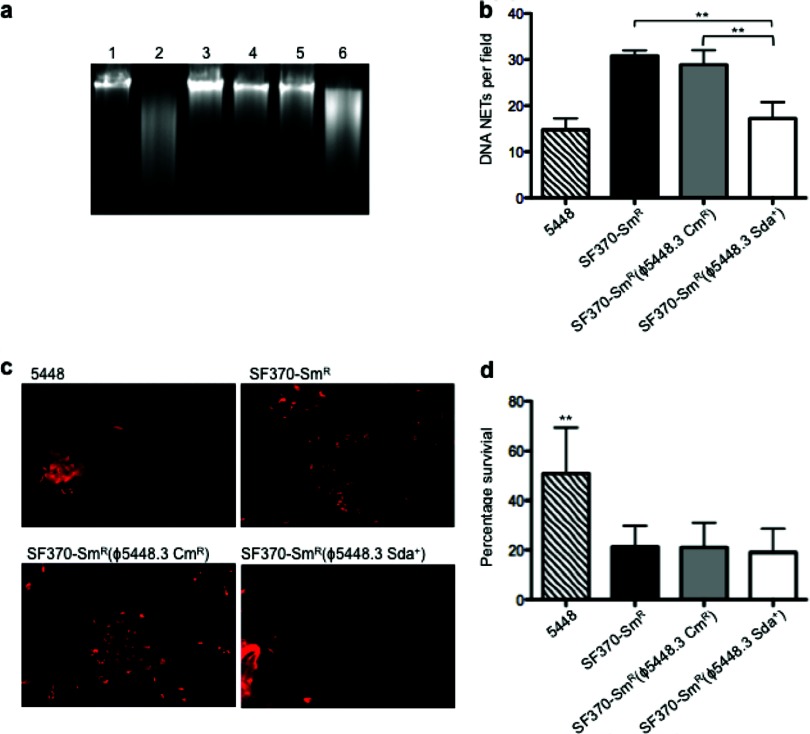
Fig 4.
Sda1 activities and resistance to neutrophil killing of lysogenized SF370 isolates. (a) DNase activities of stationary-phase GAS culture supernatants were determined by coincubation with calf thymus DNA. Lane 1, calf thymus DNA only (negative control); lane 2, 5448; lane 3, 5448Δsda; lane 4, SF370-Smr; lane 5, SF370-Smr(ϕ5448.3 Cmr); lane 6, SF370-Smr(ϕ5448.3 Sda+). Only 5448 and SF370-Smr(ϕ5448.3 Sda+) supernatants, containing Sda1, degraded DNA. (b) The bar graph shows the quantification of DNA NET induction per field of view after exposure to Sytox orange in wild-type GAS M1T1 (5448) and GAS M1 (SF370-Smr) and in the lysogens SF370-Smr(ϕ5448.3 Cmr) and SF370-Smr(ϕ5448.3 Sda+). In SF370-Smr(ϕ5448.3 Sda+), NET clearance was significantly greater than that for M1 GAS strains SF370-Smr and SF370-Smr(ϕ5448.3 Cmr) (P, <0.0007 for both comparisons) and comparable to that for the wild-type strain 5448. Values are arithmetic means plus standard errors (error bars) and are representative of three independent experiments performed in triplicate. Statistical 1-way analysis of variance with Dunnett's multiple-comparison posttest (P, <0.05) was performed using GraphPad Prism software. (c) Visualization of Sytox orange-stained DNA NETs (red) induced by coincubation of GAS serotype M1T1 strain 5448, M1 strain SF370-Smr, SF370-Smr(ϕ5448.3 Cmr), and SF370-Smr(ϕ5448.3 Sda+) with human neutrophils (MOI, 0.1). Fewer NETs can be observed for the SF370-Smr(ϕ5448.3 Sda+) lysogen, expressing Sda1, than for strains SF370-Smr and SF370-Smr(ϕ5448.3 Cmr). (d) Percentage of survival following coculture with human neutrophils in vitro. No significant difference was observed between the SF370-Smr parent strain and the lysogens, all of which were less neutrophil resistant than the M1T1 strain 5448 (P, <0.0008). Bars indicate means; error bars, standard deviations (n = 3). Statistical 1-way analysis of variance with Bonferroni's multiple-comparison posttest (P, <0.05) was performed using GraphPad Prism software.