Fig 2.
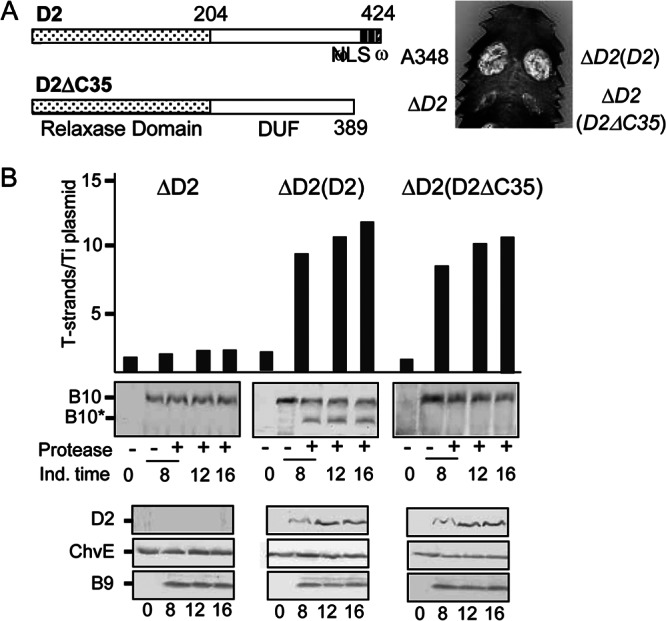
Contribution of a VirD2 C-terminal translocation signal to T-DNA transfer and VirB10 activation. (A) D2, VirD2 full length; ΔC35, C-terminal truncation mutant. Domains: relaxase, domain of unknown function (DUF), and C-terminal nuclear localization sequence (NLS), ω sequence. Numbers correspond to residues relative to the beginning of the protein. Plant leaf was inoculated with A348, wild-type strain; ΔD2 mutant, Mx355 (virD2::Tn3HoHo1 mutant) with pKA42 expressing virD4; ΔD2(D2), Mx355 with pKA250 expressing virD2 and virD4; ΔD2(D2ΔC35), Mx355 with pKA251 expressing virD2ΔC35 and virD4. (B) Numbers of T-strands per Ti plasmid in strains ΔD2, Mx355(pKA42); ΔD2(D2), Mx355(pKA250); and ΔD2(D2ΔC35), Mx355(pKA251) (see the text for details). Ind. time, T-strand levels determined at 0, 8, 12, and 16 h following vir gene induction with AS. Protease, spheroplasts were treated with S. griseus protease (+) or left untreated (−). B10, VirB10 (∼48-kDa) B10*, VirB10* proteolytic degradation product (∼40 kDa) detected by immunostaining with anti-VirB10 antibodies. Bottom, steady-state levels of D2 (VirD2), ChvE, B9 (VirB9) at the postinduction times induced as assessed by immunostaining with the respective antibodies.
