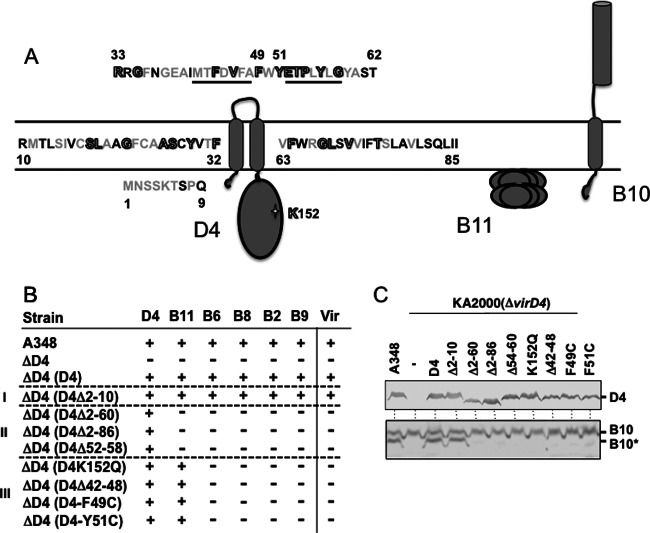
Fig 4.
Effects of VirD4 mutations on DNA transfer and VirB10 activation. (A) Schematic showing VirD4, VirB11, and VirB10 membrane topologies. The N-terminal 85 residues of VirD4 are depicted, with residues that are not conserved (black), similar (gray), or invariant (gray with black outline) among homologs associated with T4SSs in related rhizobial species. Underlined residues denote deletion mutations Δ42-48 and Δ52-58; residues F49 and Y51 were substituted with Cys, and K152 in the Walker A motif was substituted with Gln. (B) A348 (WT) or K2001 (ΔvirD4) expressing variant forms of VirD4 were assayed for T-DNA transfer through the VirB/VirD4 T4SS. Table lists: strains, VirD4 variants; D4-B9, VirD4 and VirB subunits with a detectable (+) or undetectable (−) T-DNA interaction as monitored by TrIP; Vir, T-DNA transfer to plants as monitored by virulence (+, virulent; −, avirulent). Mutant strains were grouped into 3 classes: (i) permissive, (ii) blocked T-DNA transfer from VirD4 to VirB11, (iii) blocked T-DNA transfer from VirB11 to VirB6. (C) A348 and KA2000 (ΔvirD4) strains lacking (−) or producing D4 (native VirD4) or the VirD4 mutant derivatives shown were assayed for steady-state accumulation of the VirD4 variants (top) and B10 (native VirB10) and B10* (VirB10*) upon protease treatment of spheroplasts (bottom) by immunostaining with the anti-VirD4 or -VirB10 antibodies.