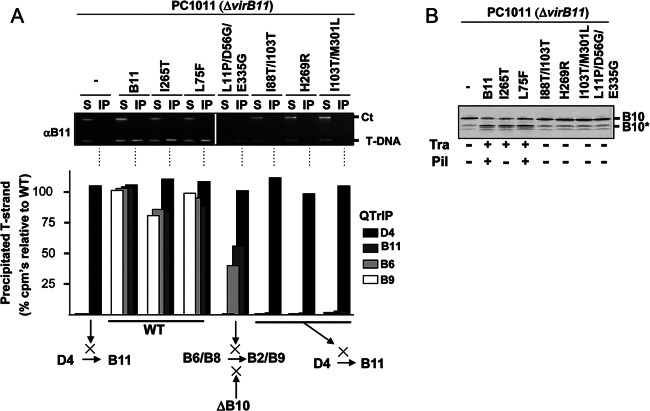
Fig 5.
Effects of VirB11 substitution mutations on T-DNA transfer and VirB10 activation. (A) Top, transfer of T-DNA to native and mutant forms of VirB11, as assessed with the TrIP assay. Strains: PC1011 (ΔvirB11) lacking (−) or producing B11 (native VirB11) or the mutant derivatives shown. S, supernatant from the IP reaction; IP, immunoprecipitated material recovered with anti-VirB11 antibodies (αB11); Ct, Ti plasmid control amplicon; T-DNA, T-DNA amplicon. Bottom, results of QTrIP assays showing levels of processed T-DNA (T-strand) recovered by precipitation with anti-VirD4, -VirB11, -VirB6, and -VirB9 antibodies relative to strain A348 (WT strain), as described previously (17). Results are presented as a percentage of the T-DNA detected in the immunoprecipitates from WT A348. Schematics summarize effects of dominant, nonfunctional mutations in blocking DNA transfer at specific stages of the translocation pathway. WT, no stage-specific block (conferred by native VirB11 and the dominant, functional mutant proteins shown). A ΔvirB10 mutation (ΔB10) blocks later-stage transfer from VirB6/VirB8 to VirB2/VirB9 (17). (B) PC1011 (ΔvirB11) lacking (−) or producing B11 (native VirB11) or the mutant derivatives shown were assayed for accumulation of B10 (native VirB10) and B10* (VirB10*) upon protease treatment of spheroplasts by immunostaining with the anti-VirB10 antibodies. Tra, T-DNA transfer to plants as monitored by virulence (+, virulent; −, avirulent,); Pil, production of detectable (+) or undetectable (−) levels of pili.