Fig 6.
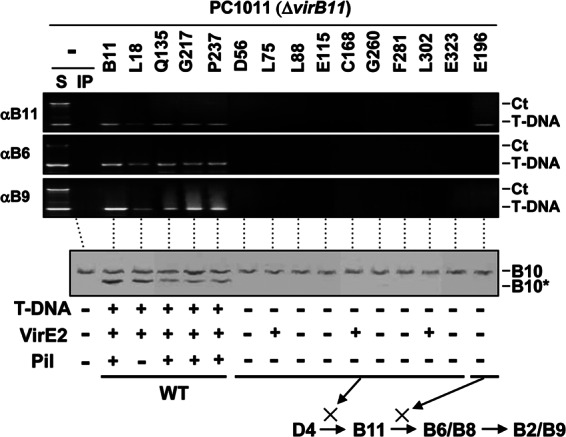
Effects of VirB11.i4 mutations on T-DNA transfer and VirB10 activation. Top, transfer of T-DNA to native and mutant forms of VirB11 and to VirB6 and VirB9, as assessed with the TrIP assay. Strains: PC1011 (ΔvirB11) lacking (−) or producing B11 (native VirB11) or the i4 mutant proteins shown. S, supernatant from the IP reaction; IP, immunoprecipitated material recovered with anti-VirB11, -VirB6, or -VirB9 antibodies (αB11, αB6, αB9). For PC1011 strains producing native or mutant forms of VirB11 (the i4 mutation is inserted after the residue listed), amplicons from the immunoprecipitate (IP) fractions are shown. Ct, control, chromosomal DNA amplicon; T-DNA, T-DNA amplicon. Bottom, PC1011 (ΔvirB11) lacking (−) or producing B11 (native VirB11) or the mutant derivatives were assayed for accumulation of B10 (native VirB10) and B10* (VirB10*) upon protease treatment of spheroplasts by immunostaining with the anti-VirB10 antibodies. T-DNA, T-DNA transfer to plants as monitored by virulence (+, virulent; −, avirulent); VirE2, transfer of VirE2 to plant cells as monitored with a mixed infection assay (+, transfer; −, no transfer). Pil, production of detectable (+) or undetectable (−) levels of pili. Schematics summarize effects of mutations in blocking DNA transfer at specific stages of the translocation pathway. WT, no stage-specific block.
