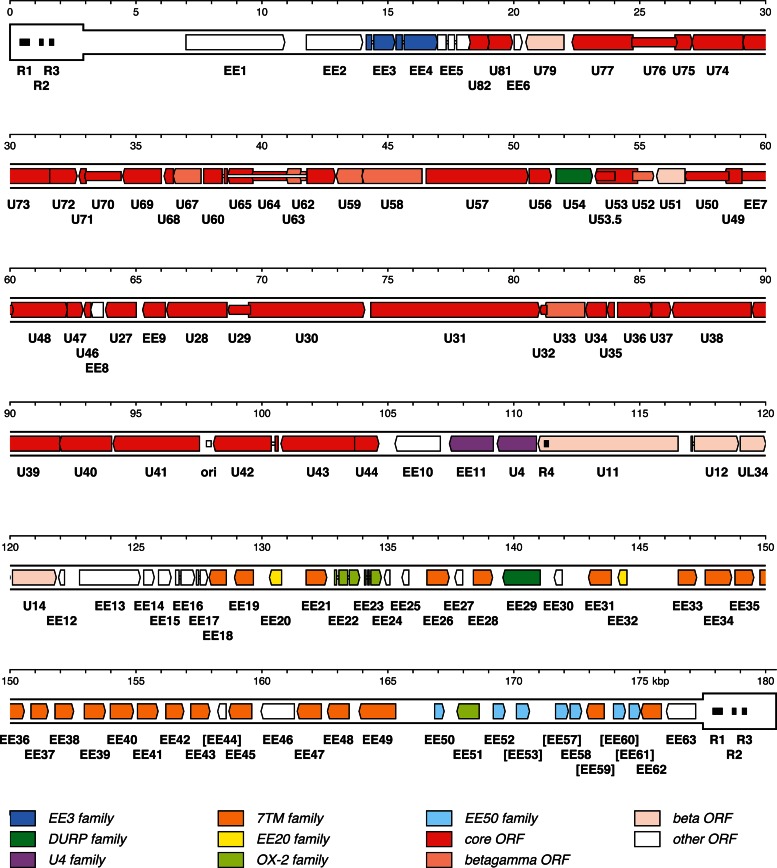
Fig 2.
Map of the EEHV1A genome. TR is shown in a thicker format than U. Predicted functional ORFs are indicated by colored arrows grouped according to the key shown at the bottom, with gene nomenclature below the ORFs. Color indicates whether an ORF has an ortholog in the ancestor of the family Herpesviridae (core ORFs), whether it is conserved among beta- and gammaherpesviruses but not alphaherpesviruses (beta-gamma ORFs), whether it is conserved among some (but not necessarily all) betaherpesviruses (beta ORFs), whether it belongs to a paralogous family (various designations), or whether it belongs to none of these categories (other ORFs). U54 is both a beta ORF and a member of the DURP family, and U4 is both a beta ORF and a member of the U4 family. Introns are shown as narrow white bars. The names of fragmented ORFs are given in square brackets, with the ORFs depicted as intact. A putative origin of DNA replication (ori) is marked by a white-shaded rectangle, and four tandem repeats longer than the read length (R1 to R4) are marked by black-shaded rectangles.