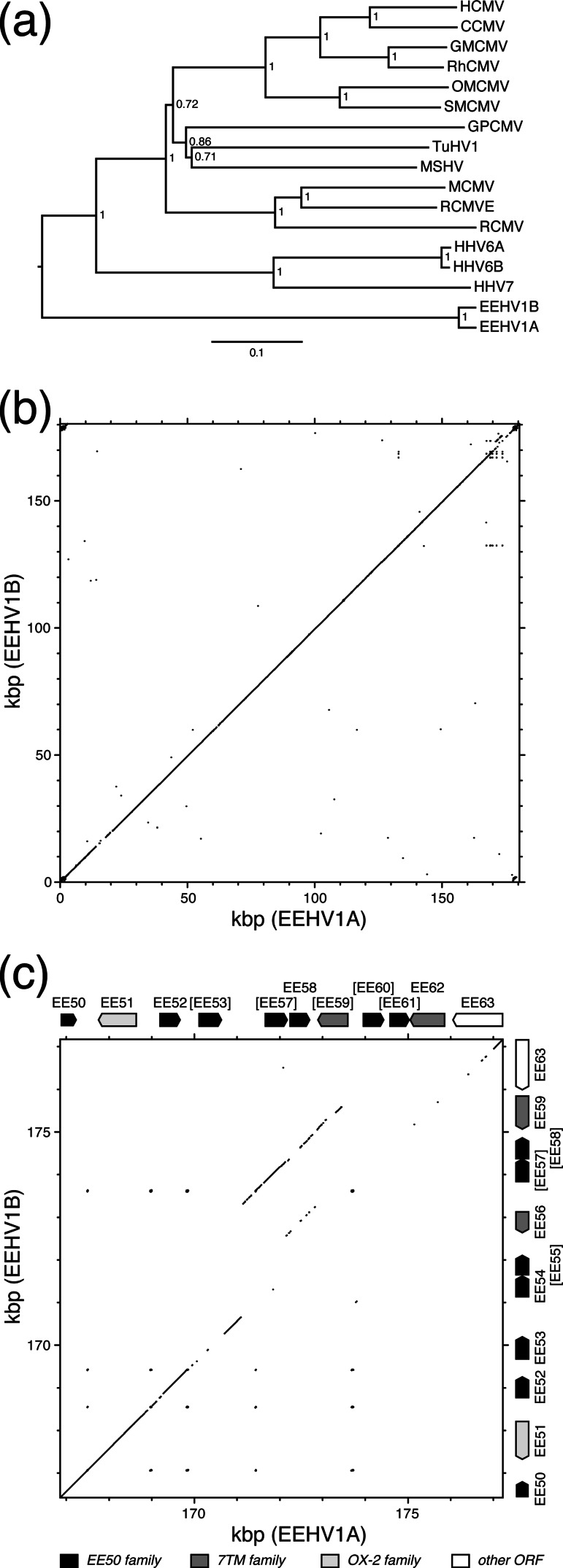
Fig 3.
Comparisons of the EEHV1A and EEHV1B genomes. (a) Phylogenetic analysis of the concatenated amino acid sequences of U38, U39, U40, U41, U57, U60, U77, and U81 from EEHV1A and EEHV1B and their orthologs in other betaherpesviruses. Abbreviations are given in the legend to Fig. 1 and also include the following: RhCMV, rhesus cytomegalovirus; TuHV1, tupaiid herpesvirus 1; MSHV, Miniopterus schreibersii herpesvirus; and RCMVE, rat cytomegalovirus England. The tree was constructed by using the neighbor-joining method, rooting at the midpoint. Confidence levels were calculated by using bootstrapping (2,000 replicates) and are shown as fractions. The scale shows nucleotide differences/nucleotide. (b) Matrix sequence comparison plot of the complete EEHV1A and EEHV1B genomes. (c) Matrix sequence comparison plot of the regions near the right terminus of the EEHV1A and EEHV1B genomes, corresponding to an expansion of part of the upper portion of the plot shown in panel b. The layout of ORFs in each sequence is illustrated, with shading indicating ORF families provided in the key at the bottom. The plots in panels b and c were computed by using GCG Compare and GCG Dotplot (window, 25; stringency, 21).