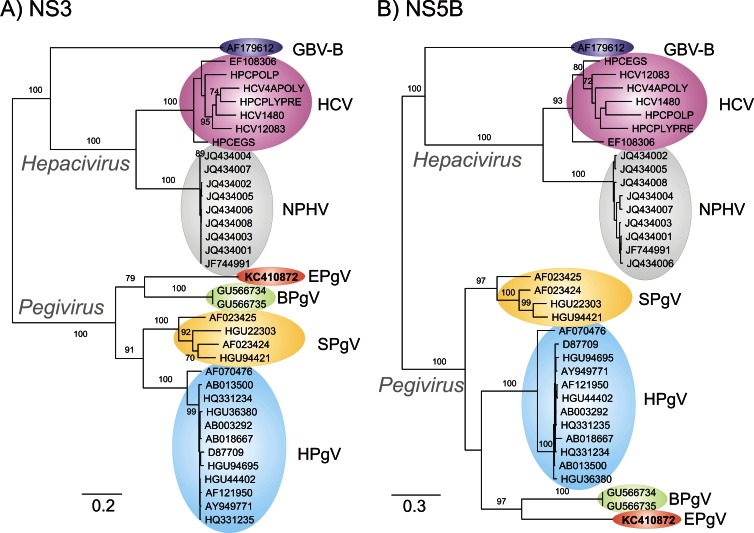
Fig 2.
Maximum-likelihood trees of amino acid sequences from the NS3 (nt 3545 to 5523 in the C35 EPgV genome) and NS5B (nt 8743 to 10231) regions of EPgV and other pegiviruses. Pegiviruses were compared with homologous regions of hepaciviruses (HCV genotypes 1a to 7a, NPHV, and GBV-B). Sequences used in phylogenetic analysis: SPgV and GBV-Ccpz, HGU22303, AF023424, AF023425, HGU94421, GVU84961, and AF070476; HPgV (all complete genomes showing >2% divergence from each other), AB008342, D87709, D87711, AB003290, D87714, D90601, D87712, D87708, D87715, D87263, D87262, AB003293, AB003288, D87710, D87713, HGU94695, HGU75356, AF006500, AB013501, HGU63715, AF309966, AF121950, AF031827, AB003289, AY196904, D87255, HGU44402, AF104403, D90600, HGU45966, AY949771, AB008336, AB003291, HGU36380, AB013500, AB021287, AB018667, AB003292, HQ331234, HQ331235, HQ331233, AB010193, AF017560, AB008335, AF081782, AF031828, and AF031829; BPgV, GU566735 and GU566734.